A mechanism for DNA-PK activation requiring unique contributions from each strand of a DNA terminus and implications for microhomology-mediated nonhomologous DNA end joining
- PMID: 18515838
- PMCID: PMC2475626
- DOI: 10.1093/nar/gkn344
A mechanism for DNA-PK activation requiring unique contributions from each strand of a DNA terminus and implications for microhomology-mediated nonhomologous DNA end joining
Abstract
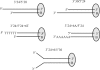
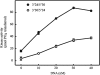
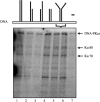
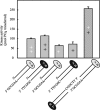
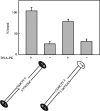
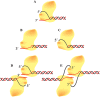
DNA-dependent protein kinase (DNA-PK) is an essential component of the nonhomologous end joining pathway (NHEJ), responsible for the repair of DNA double-strand breaks. Ku binds a DSB and recruits the catalytic subunit, DNA-PKcs, where it is activated once the kinase is bound to the DSB. The precise mechanism by which DNA activates DNA-PK remains unknown. We have investigated the effect of DNA structure on DNA-PK activation and results demonstrate that in Ku-dependent DNA-PKcs reactions, DNA-PK activation with DNA effectors containing two unannealed ends was identical to activation observed with fully duplex DNA effectors of the same length. The presence of a 6-base single-stranded extension resulted in decreased activation compared to the fully duplex DNA. DNA-PK activation using DNA effectors with compatible termini displayed increased activity compared to effectors with noncompatible termini. A strand orientation preference was observed in these reactions and suggests a model where the 3' strand of the terminus is responsible for annealing and the 5' strand is involved in activation of DNA-PK. These results demonstrate the influence of DNA structure and orientation on DNA-PK activation and provide a molecular mechanism of activation resulting from compatible termini, an essential step in microhomology-mediated NHEJ.
Figures
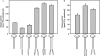
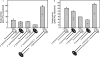
References
-
- Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. - PubMed
-
- Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. ONC. 2005;24:949–961. - PubMed
-
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

