Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region
- PMID: 18519571
- PMCID: PMC2516975
- DOI: 10.1074/jbc.M709651200
Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region
Abstract
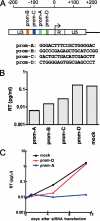
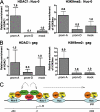
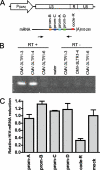
In some mammalian systems small interfering RNAs (siRNA) targeting homologous sequences in promoter regions of genes induce transcriptional gene silencing (TGS). We have previously reported the induction of TGS by an siRNA (prom-A siRNA) targeting the tandem NF-kappaB-binding motifs within the human immunodeficiency virus, type 1 (HIV-1), promoter region. Here we report that induction of TGS by prom-A siRNA is accompanied by immediate and sustained local recruitment of Argonaute-1 (Ago1), histone deacetylase-1 (HDAC1), and induction of dimethylation of histone 3 at lysine 9 (H3K9me2), processes known to be associated with transcriptional silencing. Elevated levels of H3K9me2 and HDAC1 spread upstream of the target sequence, and elevated H3K9me2 levels also spread downstream into the coding region. Moreover, this siRNA induces an immediate change in DNA accessibility to restriction enzyme digestion in the region of the transcription initiation site of the HIV-1. This change in accessibility is because of the relocation of a nucleosome known to be associated with this region of the integrated pro-virus. Although there is a theoretical possibility that the observed viral suppression could be mediated by the PTGS mechanism with this siRNA acting at the 3 (R)-long term repeat of the virus, we demonstrate that this siRNA, and three other U3 targeted siRNAs, are inefficient inducers of PTGS. These data strongly suggest that siRNA targeting the promoter region acts predominantly at a site within the 5 (R)-long term repeat of HIV to induce transcriptional silencing and alterations to chromatin structure of the HIV promoter region that extend well beyond the immediate siRNA target site. These induced changes are consistent with those described in latent HIV-1 infection.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

