Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development
- PMID: 18519645
- PMCID: PMC2418590
- DOI: 10.1101/gad.461808
Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development
Abstract
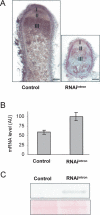
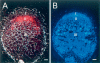
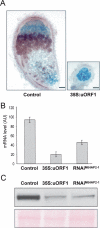
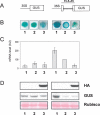
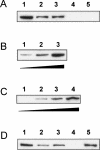
MtHAP2-1 is a CCAAT-binding transcription factor from the model legume Medicago truncatula. We previously showed that MtHAP2-1 expression is regulated both spatially and temporally by microRNA169. Here we present a novel regulatory mechanism controlling MtHAP2-1 expression. Alternative splicing of an intron in the MtHAP2-1 5'leader sequence (LS) becomes predominant during the development of root nodules, leading to the production of a small peptide, uORF1p. Our results indicate that binding of uORF1p to MtHAP2-1 5'LS mRNA leads to reduced accumulation of the MtHAP2-1 transcript and may contribute to spatial restriction of MtHAP2-1 expression within the nodule. We propose that miR169 and uORF1p play essential, sequential, and nonredundant roles in regulating MtHAP2-1 expression. Importantly, in contrast to previously described cis-acting uORFs, uORF1p is able to act in trans to down-regulate gene expression. Our work thus contributes to a better understanding of the action of upstream ORFs (uORFs) in the regulation of gene expression.
Figures

References
-
- Ardourel M., Demont N., Debelle F., Maillet F., de Billy F., Prome J.C., Denarie J., Truchet G. Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. - PMC - PubMed
-
- Ben-Naim O., Eshed R., Parnis A., Teper-Bamnolker P., Shalit A., Coupland G., Samach A., Lifschitz E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46:462–476. - PubMed
-
- Bhattacharya A., Deng J.M., Zhang Z., Behringer R., de Crombrugghe B., Maity S.N. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. - PubMed
-
- Blencowe B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
