Ibis T5000: a universal biosensor approach for microbiology
- PMID: 18521073
- PMCID: PMC7097577
- DOI: 10.1038/nrmicro1918
Ibis T5000: a universal biosensor approach for microbiology
Abstract
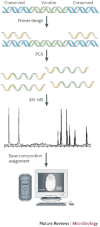
We describe a new technology, the Ibis T5000, for the identification of pathogens in clinical and environmental samples. The Ibis T5000 couples nucleic acid amplification to high-performance electrospray ionization mass spectrometry and base-composition analysis. The system enables the identification and quantification of a broad set of pathogens, including all known bacteria, all major groups of pathogenic fungi and the major families of viruses that cause disease in humans and animals, along with the detection of virulence factors and antibiotic resistance markers.
Conflict of interest statement
The authors are employees of Ibis Biosciences.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

