Yeast UCS proteins promote actomyosin interactions and limit myosin turnover in cells
- PMID: 18523008
- PMCID: PMC2430351
- DOI: 10.1073/pnas.0802874105
Yeast UCS proteins promote actomyosin interactions and limit myosin turnover in cells
Abstract
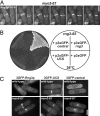
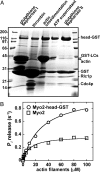
Two functions are proposed for the conserved family of UCS proteins: helping to fold myosin motor proteins and stimulating the motor function of folded myosins. We examined both functions in yeast. The fission yeast UCS protein (Rng3p) concentrates in nodes containing myosin-II (Myo2) and other proteins that condense into the cytokinetic contractile ring. Both the N-terminal (central) and C-terminal (UCS) domains of Rng3p can concentrate independently in contractile rings, but only full-length Rng3p supports contractile ring function in vivo. The presence of Rng3p in ATPase assays doubles the apparent affinity (K(ATPase)) of both native Myo2 and recombinant heads of Myo2 for actin filaments. Rng3p promotes gliding of actin filaments by full-length Myo2 molecules, but not Myo2 heads alone. Myo2 isolated from mutant strains defective for Rng3p function is soluble and supports actin filament gliding. In budding yeast the single UCS protein (She4p) acts on both myosin-I isoforms (Myo3p and Myo5p) and one of two myosin-V isoforms (Myo4p). Myo5p turns over approximately 10 times faster in she4Delta cells than wild-type cells, reducing the level of Myo5p in cells 10-fold and in cortical actin patches approximately 4-fold. Nevertheless, Myo5p isolated from she4Delta cells has wild-type ATPase and motility activities. Thus, a fraction of this yeast myosin can fold de novo in the absence of UCS proteins, but UCS proteins promote myosin stability and interactions with actin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
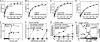



References
-
- Hutagalung AH, Landsverk ML, Price MG, Epstein HF. The UCS family of myosin chaperones. J Cell Sci. 2002;115:3983–3990. - PubMed
-
- Kachur T, Ao W, Berger J, Pilgrim D. Maternal UNC-45 is involved in cytokinesis and colocalizes with non-muscle myosin in the early Caenorhabditis elegans embryo. J Cell Sci. 2004;117:5313–5321. - PubMed
-
- Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 2002;295:669–671. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

