Aberrant CpG island methylation in acute myeloid leukemia is accentuated at relapse
- PMID: 18523155
- PMCID: PMC2515110
- DOI: 10.1182/blood-2007-11-126227
Aberrant CpG island methylation in acute myeloid leukemia is accentuated at relapse
Abstract
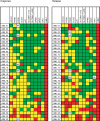
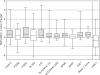
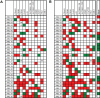
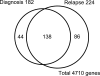
DNA methylation of CpG islands around gene transcription start sites results in gene silencing and plays a role in leukemia pathophysiology. Its impact in leukemia progression is not fully understood. We performed genomewide screening for methylated CpG islands and identified 8 genes frequently methylated in leukemia cell lines and in patients with acute myeloid leukemia (AML): NOR1, CDH13, p15, NPM2, OLIG2, PGR, HIN1, and SLC26A4. We assessed the methylation status of these genes and of the repetitive element LINE-1 in 30 patients with AML, both at diagnosis and relapse. Abnormal methylation was found in 23% to 83% of patients at diagnosis and in 47% to 93% at relapse, with CDH13 being the most frequently methylated. We observed concordance in methylation of several genes, confirming the presence of a hypermethylator pathway in AML. DNA methylation levels increased at relapse in 25 of 30 (83%) patients with AML. These changes represent much larger epigenetic dysregulation, since methylation microarray analysis of 9008 autosomal genes in 4 patients showed hypermethylation ranging from 5.9% to 13.6% (median 8.3%) genes at diagnosis and 8.0% to 15.2% (median 10.6%) genes in relapse (P < .001). Our data suggest that DNA methylation is involved in AML progression and provide a rationale for the use of epigenetic agents in remission maintenance.
Figures

References
-
- Frohling S, Scholl C, Gilliland DG, Levine RL. Genetics of myeloid malignancies: pathogenetic and clinical implications. J Clin Oncol. 2005;23:6285–6295. - PubMed
-
- Toyota M, Kopecky KJ, Toyota MO, Jair KW, Willman CL, Issa JP. Methylation profiling in acute myeloid leukemia. Blood. 2001;97:2823–2829. - PubMed
-
- Galm O, Wilop S, Luders C, et al. Clinical implications of aberrant DNA methylation patterns in acute myelogenous leukemia. Ann Hematol. 2005;84(suppl 13):39–46. - PubMed
-
- Issa JP. Methylation and prognosis: of molecular clocks and hypermethylator phenotypes. Clin Cancer Res. 2003;9:2879–2881. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

