Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B)
- PMID: 18523274
- PMCID: PMC2493419
- DOI: 10.4049/jimmunol.180.12.8102
Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B)
Abstract
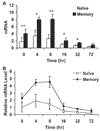
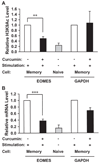
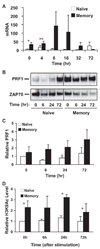
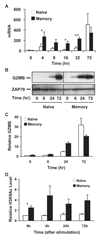
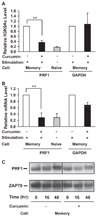
To understand the mechanism regulating the effector function of memory CD8 T cells, we examined expression and chromatin state of a key transcription factor (eomesodermin, EOMES) and two of its targets: perforin (PRF1) and granzyme B (GZMB). Accessible chromatin associated histone 3 lysine 9 acetylation (H3K9Ac) was found significantly higher at the proximal promoter and the first exon region of all three genes in memory CD8 T cells than in naive CD8 T cells. Correspondingly, EOMES and PRF1 were constitutively higher expressed in memory CD8 T cells than in naive CD8 T cells at resting and activated states. In contrast, higher expression of GZMB was induced in memory CD8 T cells than in naive CD8 T cells only after activation. Regardless of their constitutive or inducible expression, decreased H3K9Ac levels after treatment with a histone acetyltransferase inhibitor (Curcumin) led to decreased expression of all three genes in activated memory CD8 T cells. These findings suggest that H3K9Ac associated accessible chromatin state serves as a corner stone for the differentially high expression of these effector genes in memory CD8 T cells. Thus, epigenetic changes mediated via histone acetylation may provide a chromatin "memory" for the rapid and robust transcriptional response of memory CD8 T cells.
Conflict of interest statement
The authors have no financial conflict of interest.
Figures



References
-
- Dutton RW, Bradley LM, Swain SL. T cell memory. Annu. Rev. Immunol. 1998;16:201–223. - PubMed
-
- Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2002;2:251–262. - PubMed
-
- Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu. Rev. Immunol. 2007;25:171–192. - PubMed
-
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. - PubMed
-
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

