Identification of a second CtBP binding site in adenovirus type 5 E1A conserved region 3
- PMID: 18524818
- PMCID: PMC2519622
- DOI: 10.1128/JVI.00248-08
Identification of a second CtBP binding site in adenovirus type 5 E1A conserved region 3
Abstract
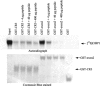
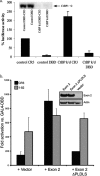
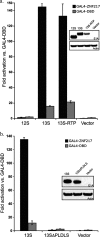
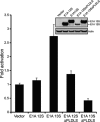
C-terminal binding protein (CtBP) binds to adenovirus early region 1A (AdE1A) through a highly conserved PXDLS motif close to the C terminus. We now have demonstrated that CtBP1 also interacts directly with the transcriptional activation domain (conserved region 3 [CR3]) of adenovirus type 5 E1A (Ad5E1A) and requires the integrity of the entire CR3 region for optimal binding. The interaction appears to be at least partially mediated through a sequence ((161)RRNTGDP(167)) very similar to a recently characterized novel CtBP binding motif in ZNF217 as well as other regions of CR3. Using reporter assays, we further demonstrated that CtBP1 represses Ad5E1A CR3-dependent transcriptional activation. Ad5E1A also appears to be recruited to the E-cadherin promoter through its interaction with CtBP. Significantly, Ad5E1A, CtBP1, and ZNF217 form a stable complex which requires CR3 and the PLDLS motif. It has been shown that Ad513SE1A, containing the CR3 region, is able to overcome the transcriptional repressor activity of a ZNF217 polypeptide fragment in a GAL4 reporter assay through recruitment of CtBP1. These results suggest a hitherto-unsuspected complexity in the association of Ad5E1A with CtBP, with the interaction resulting in transcriptional activation by recruitment of CR3-bound factors to CtBP1-containing complexes.
Figures



References
-
- Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 37481-84. - PubMed
-
- Avvakumov, N., A. E. Kajon, R. C. Hoeben, and J. S. Mymryk. 2004. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology 329477-492. - PubMed
-
- Balasubramanian, P., L. J. Zhao, and G. Subramanian. 2003. Nicotinanide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 537157-160. - PubMed
-
- Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control and cell biology from adenovirus. Oncogene 247673-7685. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

