The evolution of the DLK1-DIO3 imprinted domain in mammals
- PMID: 18532878
- PMCID: PMC2408620
- DOI: 10.1371/journal.pbio.0060135
The evolution of the DLK1-DIO3 imprinted domain in mammals
Abstract
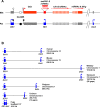
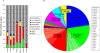
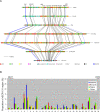
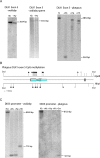
A comprehensive, domain-wide comparative analysis of genomic imprinting between mammals that imprint and those that do not can provide valuable information about how and why imprinting evolved. The imprinting status, DNA methylation, and genomic landscape of the Dlk1-Dio3 cluster were determined in eutherian, metatherian, and prototherian mammals including tammar wallaby and platypus. Imprinting across the whole domain evolved after the divergence of eutherian from marsupial mammals and in eutherians is under strong purifying selection. The marsupial locus at 1.6 megabases, is double that of eutherians due to the accumulation of LINE repeats. Comparative sequence analysis of the domain in seven vertebrates determined evolutionary conserved regions common to particular sub-groups and to all vertebrates. The emergence of Dlk1-Dio3 imprinting in eutherians has occurred on the maternally inherited chromosome and is associated with region-specific resistance to expansion by repetitive elements and the local introduction of noncoding transcripts including microRNAs and C/D small nucleolar RNAs. A recent mammal-specific retrotransposition event led to the formation of a completely new gene only in the eutherian domain, which may have driven imprinting at the cluster.
Conflict of interest statement
Figures



References
-
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. - PubMed
-
- Constancia M, Kelsey G, Reik W. Resourceful imprinting. Nature. 2004;432:53–57. - PubMed
-
- Kaneko-Ishino T, Kohda T, Ono R, Ishino F. Complementation hypothesis: the necessity of a monoallelic gene expression mechanism in mammalian development. Cytogenet Genome Res. 2006;113:24–30. - PubMed
-
- Charalambous M, Ferguson-Smith AC, Da Rocha ST. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrin Diabetes, Obesity. 2007;14:3–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

