Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor
- PMID: 18533182
- PMCID: PMC2576750
- DOI: 10.1016/j.jmb.2008.04.061
Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor
Abstract
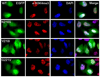
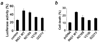
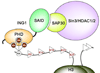
Inhibitor of growth 1 (ING1) is implicated in oncogenesis, DNA damage repair, and apoptosis. Mutations within the ING1 gene and altered expression levels of ING1 are found in multiple human cancers. Here, we show that both DNA repair and apoptotic activities of ING1 require the interaction of the C-terminal plant homeodomain (PHD) finger with histone H3 trimethylated at Lys4 (H3K4me3). The ING1 PHD finger recognizes methylated H3K4 but not other histone modifications as revealed by the peptide microarrays. The molecular mechanism of the histone recognition is elucidated based on a 2.1 A-resolution crystal structure of the PHD-H3K4me3 complex. The K4me3 occupies a deep hydrophobic pocket formed by the conserved Y212 and W235 residues that make cation-pi contacts with the trimethylammonium group. Both aromatic residues are essential in the H3K4me3 recognition, as substitution of these residues with Ala disrupts the interaction. Unlike the wild-type ING1, the W235A mutant, overexpressed in the stable clones of melanoma cells or in HT1080 cells, was unable to stimulate DNA repair after UV irradiation or promote DNA-damage-induced apoptosis, indicating that H3K4me3 binding is necessary for these biological functions of ING1. Furthermore, N216S, V218I, and G221V mutations, found in human malignancies, impair the ability of ING1 to associate with H3K4me3 or to induce nucleotide repair and cell death, linking the tumorigenic activity of ING1 with epigenetic regulation. Together, our findings reveal the critical role of the H3K4me3 interaction in mediating cellular responses to genotoxic stresses and offer new insight into the molecular mechanism underlying the tumor suppressive activity of ING1.
Figures



Similar articles
-
Structural insight into histone recognition by the ING PHD fingers.Curr Drug Targets. 2009 May;10(5):432-41. doi: 10.2174/138945009788185040. Curr Drug Targets. 2009. PMID: 19442115 Free PMC article. Review.
-
Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2.Nature. 2006 Jul 6;442(7098):100-3. doi: 10.1038/nature04814. Epub 2006 May 21. Nature. 2006. PMID: 16728977 Free PMC article.
-
Ing1 functions in DNA demethylation by directing Gadd45a to H3K4me3.Genes Dev. 2013 Feb 1;27(3):261-73. doi: 10.1101/gad.186916.112. Genes Dev. 2013. PMID: 23388825 Free PMC article.
-
ING1 and ING2: multifaceted tumor suppressor genes.Cell Mol Life Sci. 2013 Oct;70(20):3753-72. doi: 10.1007/s00018-013-1270-z. Epub 2013 Feb 15. Cell Mol Life Sci. 2013. PMID: 23412501 Free PMC article. Review.
-
Mechanism of Histone H3K4me3 Recognition by the Plant Homeodomain of Inhibitor of Growth 3.J Biol Chem. 2016 Aug 26;291(35):18326-41. doi: 10.1074/jbc.M115.690651. Epub 2016 Jun 8. J Biol Chem. 2016. PMID: 27281824 Free PMC article.
Cited by
-
Loss of the N-terminal methyltransferase NRMT1 increases sensitivity to DNA damage and promotes mammary oncogenesis.Oncotarget. 2015 May 20;6(14):12248-63. doi: 10.18632/oncotarget.3653. Oncotarget. 2015. PMID: 25909287 Free PMC article.
-
ING Proteins: Tumour Suppressors or Oncoproteins.Cancers (Basel). 2021 Apr 27;13(9):2110. doi: 10.3390/cancers13092110. Cancers (Basel). 2021. PMID: 33925563 Free PMC article. Review.
-
Reviewing the current classification of inhibitor of growth family proteins.Cancer Sci. 2009 Jul;100(7):1173-9. doi: 10.1111/j.1349-7006.2009.01183.x. Epub 2009 Apr 28. Cancer Sci. 2009. PMID: 19432890 Free PMC article. Review.
-
Impaired DNA demethylation of C/EBP sites causes premature aging.Genes Dev. 2018 Jun 1;32(11-12):742-762. doi: 10.1101/gad.311969.118. Epub 2018 Jun 8. Genes Dev. 2018. PMID: 29884649 Free PMC article.
-
Readout of histone methylation by Trim24 locally restricts chromatin opening by p53.Nat Struct Mol Biol. 2023 Jul;30(7):948-957. doi: 10.1038/s41594-023-01021-8. Epub 2023 Jun 29. Nat Struct Mol Biol. 2023. PMID: 37386214 Free PMC article.
References
-
- Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–420. - PubMed
-
- Helbing CC, Veillette C, Riabowol K, Johnston RN, Garkavtsev I. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res. 1997;57:1255–1258. - PubMed
-
- Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature. 1998;391:295–298. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

