An analytic and systematic framework for estimating metabolic flux ratios from 13C tracer experiments
- PMID: 18534038
- PMCID: PMC2430715
- DOI: 10.1186/1471-2105-9-266
An analytic and systematic framework for estimating metabolic flux ratios from 13C tracer experiments
Abstract
Background: Metabolic fluxes provide invaluable insight on the integrated response of a cell to environmental stimuli or genetic modifications. Current computational methods for estimating the metabolic fluxes from 13C isotopomer measurement data rely either on manual derivation of analytic equations constraining the fluxes or on the numerical solution of a highly nonlinear system of isotopomer balance equations. In the first approach, analytic equations have to be tediously derived for each organism, substrate or labelling pattern, while in the second approach, the global nature of an optimum solution is difficult to prove and comprehensive measurements of external fluxes to augment the 13C isotopomer data are typically needed.
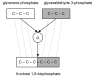
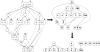
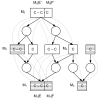
Results: We present a novel analytic framework for estimating metabolic flux ratios in the cell from 13C isotopomer measurement data. In the presented framework, equation systems constraining the fluxes are derived automatically from the model of the metabolism of an organism. The framework is designed to be applicable with all metabolic network topologies, 13C isotopomer measurement techniques, substrates and substrate labelling patterns. By analyzing nuclear magnetic resonance (NMR) and mass spectrometry (MS) measurement data obtained from the experiments on glucose with the model micro-organisms Bacillus subtilis and Saccharomyces cerevisiae we show that our framework is able to automatically produce the flux ratios discovered so far by the domain experts with tedious manual analysis. Furthermore, we show by in silico calculability analysis that our framework can rapidly produce flux ratio equations--as well as predict when the flux ratios are unobtainable by linear means--also for substrates not related to glucose.
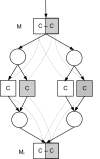
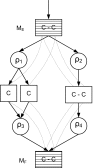
Conclusion: The core of 13C metabolic flux analysis framework introduced in this article constitutes of flow and independence analysis of metabolic fragments and techniques for manipulating isotopomer measurements with vector space techniques. These methods facilitate efficient, analytic computation of the ratios between the fluxes of pathways that converge to a common junction metabolite. The framework can been seen as a generalization and formalization of existing tradition for computing metabolic flux ratios where equations constraining flux ratios are manually derived, usually without explicitly showing the formal proofs of the validity of the equations.
Figures



Similar articles
-
Designer labels for plant metabolism: statistical design of isotope labeling experiments for improved quantification of flux in complex plant metabolic networks.Mol Biosyst. 2013 Jan 27;9(1):99-112. doi: 10.1039/c2mb25253h. Epub 2012 Nov 1. Mol Biosyst. 2013. PMID: 23114423
-
Theoretical aspects of 13C metabolic flux analysis with sole quantification of carbon dioxide labeling.Comput Biol Chem. 2005 Apr;29(2):121-33. doi: 10.1016/j.compbiolchem.2005.02.005. Comput Biol Chem. 2005. PMID: 15833440
-
Bidirectional reaction steps in metabolic networks: IV. Optimal design of isotopomer labeling experiments.Biotechnol Bioeng. 1999;66(2):86-103. doi: 10.1002/(sici)1097-0290(1999)66:2<86::aid-bit2>3.0.co;2-a. Biotechnol Bioeng. 1999. PMID: 10567067
-
In vivo stationary flux analysis by 13C labeling experiments.Adv Biochem Eng Biotechnol. 1996;54:109-54. doi: 10.1007/BFb0102334. Adv Biochem Eng Biotechnol. 1996. PMID: 8623613 Review.
-
Application of stable isotopes and mass isotopomer distribution analysis to the study of intermediary metabolism of nutrients.J Anim Sci. 2006 Apr;84 Suppl:E50-9. doi: 10.2527/2006.8413_supple50x. J Anim Sci. 2006. PMID: 16582092 Review.
Cited by
-
OpenFLUX: efficient modelling software for 13C-based metabolic flux analysis.Microb Cell Fact. 2009 May 1;8:25. doi: 10.1186/1475-2859-8-25. Microb Cell Fact. 2009. PMID: 19409084 Free PMC article.
-
13C metabolic flux analysis: Classification and characterization from the perspective of mathematical modeling and application in physiological research of neural cell.Front Mol Neurosci. 2022 Sep 8;15:883466. doi: 10.3389/fnmol.2022.883466. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36157075 Free PMC article. Review.
-
Comparative genome-scale reconstruction of gapless metabolic networks for present and ancestral species.PLoS Comput Biol. 2014 Feb 6;10(2):e1003465. doi: 10.1371/journal.pcbi.1003465. eCollection 2014 Feb. PLoS Comput Biol. 2014. PMID: 24516375 Free PMC article.
-
Software LS-MIDA for efficient mass isotopomer distribution analysis in metabolic modelling.BMC Bioinformatics. 2013 Jul 9;14:218. doi: 10.1186/1471-2105-14-218. BMC Bioinformatics. 2013. PMID: 23837681 Free PMC article.
-
Integrating quantitative proteomics and metabolomics with a genome-scale metabolic network model.Bioinformatics. 2010 Jun 15;26(12):i255-60. doi: 10.1093/bioinformatics/btq183. Bioinformatics. 2010. PMID: 20529914 Free PMC article.
References
-
- Varma A, Palsson B. Metabolic flux balancing: basic concepts, scientific and practical use. Nature Biotechnology. 1994;12:994–998. doi: 10.1038/nbt1094-994. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

