How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan)
- PMID: 18535144
- PMCID: PMC2415748
- DOI: 10.1128/MMBR.00027-07
How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan)
Abstract
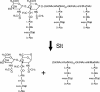
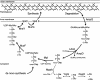
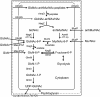
The phenomenon of peptidoglycan recycling is reviewed. Gram-negative bacteria such as Escherichia coli break down and reuse over 60% of the peptidoglycan of their side wall each generation. Recycling of newly made peptidoglycan during septum synthesis occurs at an even faster rate. Nine enzymes, one permease, and one periplasmic binding protein in E. coli that appear to have as their sole function the recovery of degradation products from peptidoglycan, thereby making them available for the cell to resynthesize more peptidoglycan or to use as an energy source, have been identified. It is shown that all of the amino acids and amino sugars of peptidoglycan are recycled. The discovery and properties of the individual proteins and the pathways involved are presented. In addition, the possible role of various peptidoglycan degradation products in the induction of beta-lactamase is discussed.
Figures





References
-
- Adam, A., and E. Lederer. 1984. Muramyl peptides: immunomodulators, sleep factors, and vitamins. Med. Res. Rev. 4111-152. - PubMed
-
- Arminjon, F., M. Guinand, M. J. Vacheron, and G. Michel. 1977. Specificity profiles of the membrane-bound γ-D-glutamyl-(L)meso-diaminopimelateendopeptidase and LD-carboxypeptidase from Bacillus sphaericus 9602. Eur. J. Biochem. 73557-565. - PubMed
-
- Asensio, C., and M. Ruiz-Amil. 1966. N-Acetyl-D-glucosamine kinase. II. Escherichia coli. Methods Enzymol. 9421-425.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

