Structural biology of pectin degradation by Enterobacteriaceae
- PMID: 18535148
- PMCID: PMC2415742
- DOI: 10.1128/MMBR.00038-07
Structural biology of pectin degradation by Enterobacteriaceae
Abstract
Pectin is a structural polysaccharide that is integral for the stability of plant cell walls. During soft rot infection, secreted virulence factors from pectinolytic bacteria such as Erwinia spp. degrade pectin, resulting in characteristic plant cell necrosis and tissue maceration. Catabolism of pectin and its breakdown products by pectinolytic bacteria occurs within distinct cellular environments. This process initiates outside the cell, continues within the periplasmic space, and culminates in the cytoplasm. Although pectin utilization is well understood at the genetic and biochemical levels, an inclusive structural description of pectinases and pectin binding proteins by both extracellular and periplasmic enzymes has been lacking, especially following the recent characterization of several periplasmic components and protein-oligogalacturonide complexes. Here we provide a comprehensive analysis of the protein folds and mechanisms of pectate lyases, polygalacturonases, and carbohydrate esterases and the binding specificities of two periplasmic pectic binding proteins from Enterobacteriaceae. This review provides a structural understanding of the molecular determinants of pectin utilization and the mechanisms driving catabolite selectivity and flow through the pathway.
Figures
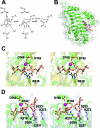
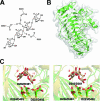



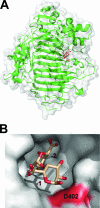
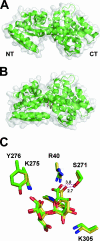

References
-
- Abbott, D. W., and A. B. Boraston. 2007. Specific recognition of saturated and 4,5-unsaturated hexuronate sugars by a periplasmic binding protein involved in pectin catabolism. J. Mol. Biol. 369759-770. - PubMed
-
- Abbott, D. W., and A. B. Boraston. 2007. The structural basis for exopolygalacturonase activity in a family 28 glycoside hydrolase. J. Mol. Biol. 3681215-1222. - PubMed
-
- Abbott, D. W., and A. B. Boraston. 2007. A family 2 pectate lyase displays a rare fold and transition metal-assisted beta-elimination. J. Biol. Chem. 28235328-35336. - PubMed
-
- Abbott, D. W., J. M. Eirin-Lopez, and A. B. Boraston. 2008. Insight into ligand diversity and novel biological roles for family 32 carbohydrate-binding modules. Mol. Biol. Evol. 25155-167. - PubMed
-
- Abbott, D. W., S. Hrynuik, and A. B. Boraston. 2007. Identification and characterization of a novel periplasmic polygalacturonic acid binding protein from Yersinia enterocolitica. J. Mol. Biol. 3671023-1033. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

