Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia
- PMID: 18535205
- PMCID: PMC2518874
- DOI: 10.1182/blood-2008-02-140012
Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia
Abstract
Diamond-Blackfan anemia (DBA) is an inherited bone marrow failure syndrome characterized by anemia, congenital abnormalities, and cancer predisposition. Small ribosomal subunit genes RPS19, RPS24, and RPS17 are mutated in approximately one-third of patients. We used a candidate gene strategy combining high-resolution genomic mapping and gene expression microarray in the analysis of 2 DBA patients with chromosome 3q deletions to identify RPL35A as a potential DBA gene. Sequence analysis of a cohort of DBA probands confirmed involvement RPL35A in DBA. shRNA inhibition shows that Rpl35a is essential for maturation of 28S and 5.8S rRNAs, 60S subunit biogenesis, normal proliferation, and cell survival. Analysis of pre-rRNA processing in primary DBA lymphoblastoid cell lines demonstrated similar alterations of large ribosomal subunit rRNA in both RPL35A-mutated and some RPL35A wild-type patients, suggesting additional large ribosomal subunit gene defects are likely present in some cases of DBA. These data demonstrate that alterations of large ribosomal subunit proteins cause DBA and support the hypothesis that DBA is primarily the result of altered ribosomal function. The results also establish that haploinsufficiency of large ribosomal subunit proteins contributes to bone marrow failure and potentially cancer predisposition.
Figures


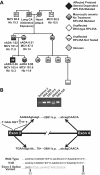

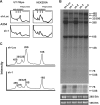

 , 5.8S; □, 28S. External and internal transcribed spacers are indicated as lines between the mature species and labeled above the primary pre-rRNA transcript. Cleavage sites, as originally proposed by Hadjiolova et al, are shown by numbered arrows above the 45S and 45S′ transcripts. The sequence of cleavage of the 45S′ pre-RNA at sites 1 and 2 results in 2 alternative processing pathways. Two additional human cleavage sites (2b and 4a) shown as numbered arrows below the transcript are inferred from these studies. The presence of a 7S precursor to 5.8S rRNA implies an additional cleavage (4a) within ITS2. An additional cleavage site corresponding to the yeast A3 site (2b) within ITS1 is also proposed. The positions of oligonucleotide probes used for Northern analysis are shown in gray below the primary transcript. (B,C) Northern analysis of rRNA from RPL35A knock-down in UT7-Epo demonstrates steady-state increases in 45S:32S and 32S:12S ratios, indicating a disruption of 32S pre-rRNA maturation with resultant decreases in mature 28S (B) and 5.8S (C) rRNA. The rRNA species are indicated to the right of each panel; the probe used is indicated in gray to the left of each panel. Eth indicates ethidium bromide; Sh-Luc, Luciferase-control transduced cells; sh-1, -2, -3, or -4, respective RPL35A shRNAs.
, 5.8S; □, 28S. External and internal transcribed spacers are indicated as lines between the mature species and labeled above the primary pre-rRNA transcript. Cleavage sites, as originally proposed by Hadjiolova et al, are shown by numbered arrows above the 45S and 45S′ transcripts. The sequence of cleavage of the 45S′ pre-RNA at sites 1 and 2 results in 2 alternative processing pathways. Two additional human cleavage sites (2b and 4a) shown as numbered arrows below the transcript are inferred from these studies. The presence of a 7S precursor to 5.8S rRNA implies an additional cleavage (4a) within ITS2. An additional cleavage site corresponding to the yeast A3 site (2b) within ITS1 is also proposed. The positions of oligonucleotide probes used for Northern analysis are shown in gray below the primary transcript. (B,C) Northern analysis of rRNA from RPL35A knock-down in UT7-Epo demonstrates steady-state increases in 45S:32S and 32S:12S ratios, indicating a disruption of 32S pre-rRNA maturation with resultant decreases in mature 28S (B) and 5.8S (C) rRNA. The rRNA species are indicated to the right of each panel; the probe used is indicated in gray to the left of each panel. Eth indicates ethidium bromide; Sh-Luc, Luciferase-control transduced cells; sh-1, -2, -3, or -4, respective RPL35A shRNAs.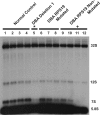
Comment in
-
Diamond-Blackfan anemia: a new facet.Blood. 2008 Sep 1;112(5):1552-3. doi: 10.1182/blood-2008-07-164202. Blood. 2008. PMID: 18725571 No abstract available.
References
-
- National Center for Biotechnology Information and McKusick-Nathans Institute of Genetic Medicine. OMIM: Johns Hopkins University; Online Mendelian inheritance in man. http://www.ncbi.nlm.nih.gov/omim.
-
- Perdahl EB, Naprstek BL, Wallace WC, Lipton JM. Erythroid failure in Diamond-Blackfan anemia is characterized by apoptosis. Blood. 1994;83:645–650. - PubMed
-
- Alter BP. Fetal erythropoiesis in stress hematopoiesis. Exp Hematol. 1979;7(suppl 5):200–209. - PubMed
-
- Glader BE, Backer K. Elevated red cell adenosine deaminase activity: a marker of disordered erythropoiesis in Diamond-Blackfan anaemia and other haematologic diseases. Br J Haematol. 1988;68:165–168. - PubMed
-
- Vlachos A, Klein GW, Lipton JM. The Diamond Blackfan Anemia Registry: tool for investigating the epidemiology and biology of Diamond-Blackfan anemia. J Pediatr Hematol Oncol. 2001;23:377–382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

