Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy
- PMID: 18535242
- PMCID: PMC2916659
- DOI: 10.1126/science.1156947
Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy
Abstract
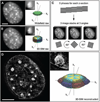
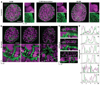
Fluorescence light microscopy allows multicolor visualization of cellular components with high specificity, but its utility has until recently been constrained by the intrinsic limit of spatial resolution. We applied three-dimensional structured illumination microscopy (3D-SIM) to circumvent this limit and to study the mammalian nucleus. By simultaneously imaging chromatin, nuclear lamina, and the nuclear pore complex (NPC), we observed several features that escape detection by conventional microscopy. We could resolve single NPCs that colocalized with channels in the lamin network and peripheral heterochromatin. We could differentially localize distinct NPC components and detect double-layered invaginations of the nuclear envelope in prophase as previously seen only by electron microscopy. Multicolor 3D-SIM opens new and facile possibilities to analyze subcellular structures beyond the diffraction limit of the emitted light.
Figures


References
-
- Born M, Wolf E, editors. Principle of Optics. Cambridge: Cambridge Univ. Press; 1998.
-
- Pawley JB, editor. Handbook of Biological Confocal Microscopy. ed. 3. New York: Springer; 2006.
-
- Abbe E. Arch. Mikrosk. Anat. 1873;9:413.
-
- Gustafsson MG, et al. Biophys. J. in press; published online 7 March 2008 (10.1529/biophysj.107.120345)
-
- Gustafsson MG. J. Microsc. 2000;198:82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

