Proteolytic processing of cGMP-dependent protein kinase I mediates nuclear cGMP signaling in vascular smooth muscle cells
- PMID: 18535260
- PMCID: PMC2749497
- DOI: 10.1161/CIRCRESAHA.108.176321
Proteolytic processing of cGMP-dependent protein kinase I mediates nuclear cGMP signaling in vascular smooth muscle cells
Abstract
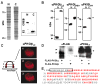
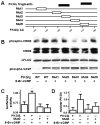
Cyclic GMP modulates gene expression in vascular smooth muscle cells (SMCs) in part by stimulating cGMP-dependent protein kinase I (PKGI) and the phosphorylation of transcription factors. In some cells, cGMP increases nuclear translocation of PKGI and PKGI-dependent phosphorylation of transcription regulators; however, these observations have been variable, and the mechanisms mediating nuclear PKGI translocation are incompletely understood. We tested the hypothesis that proteolytic cleavage of PKGI is required for cGMP-stimulated nuclear compartmentation of PKGI and phosphorylation of transcription factors. We detected an NH(2)-terminal PKGI fragment with leucine zipper domain immunoreactivity in the cytosol and endoplasmic reticulum of SMCs, but only a COOH-terminal PKGI fragment containing the catalytic region (now termed PKGIgamma) was observed in the Golgi apparatus (GA) and nucleoplasm. Posttranslational PKGI processing in the GA was critical for nuclear compartmentation of PKGIgamma because GA disruption with nocodazol or brefeldin A inhibited PKGIgamma nuclear localization. PKGIgamma immunoreactivity was particularly abundant in the nucleolus of interphase SMCs where its colocalization with the nucleolar dense fibrillar component protein fibrillarin closely matched the level of nucleolar assembly. Purified nucleolar PKGIgamma enzyme activity was insensitive to cGMP stimulation, which is consistent with its lack of the NH(2)-terminal autoinhibitory domain. Mutation of a putative proteolytic cleavage region in PKGI inhibited cGMP-mediated phosphorylation of cAMP response element-binding protein, cAMP response element-dependent transcription, and nuclear localization of PKGIgamma. These observations suggest that posttranslational modification of PKGI critically influences the nuclear translocation of PKGI and activities of cGMP in SMCs.
Conflict of interest statement
Disclosures: None.
Figures




Similar articles
-
cGMP-dependent protein kinase I gamma encodes a nuclear localization signal that regulates nuclear compartmentation and function.Cell Signal. 2014 Dec;26(12):2633-44. doi: 10.1016/j.cellsig.2014.08.004. Epub 2014 Aug 27. Cell Signal. 2014. PMID: 25172423 Free PMC article.
-
The Golgi apparatus regulates cGMP-dependent protein kinase I compartmentation and proteolysis.Am J Physiol Cell Physiol. 2015 Jun 1;308(11):C944-58. doi: 10.1152/ajpcell.00199.2014. Epub 2015 Apr 8. Am J Physiol Cell Physiol. 2015. PMID: 25855081 Free PMC article.
-
Proprotein convertases play an important role in regulating PKGI endoproteolytic cleavage and nuclear transport.Am J Physiol Lung Cell Mol Physiol. 2013 Jul 15;305(2):L130-40. doi: 10.1152/ajplung.00391.2012. Epub 2013 May 17. Am J Physiol Lung Cell Mol Physiol. 2013. PMID: 23686857 Free PMC article.
-
Regulation of vascular smooth muscle cell phenotype by cyclic GMP and cyclic GMP-dependent protein kinase.Front Biosci. 2006 Jan 1;11:356-67. doi: 10.2741/1803. Front Biosci. 2006. PMID: 16146737 Review.
-
Regulation of contractile activity in vascular smooth muscle by protein kinases.Rev Clin Basic Pharm. 1985 Jul-Dec;5(3-4):341-95. Rev Clin Basic Pharm. 1985. PMID: 3029813 Review.
Cited by
-
cGMP-dependent protein kinase I gamma encodes a nuclear localization signal that regulates nuclear compartmentation and function.Cell Signal. 2014 Dec;26(12):2633-44. doi: 10.1016/j.cellsig.2014.08.004. Epub 2014 Aug 27. Cell Signal. 2014. PMID: 25172423 Free PMC article.
-
The Golgi apparatus regulates cGMP-dependent protein kinase I compartmentation and proteolysis.Am J Physiol Cell Physiol. 2015 Jun 1;308(11):C944-58. doi: 10.1152/ajpcell.00199.2014. Epub 2015 Apr 8. Am J Physiol Cell Physiol. 2015. PMID: 25855081 Free PMC article.
-
Transforming growth factor-beta modulates the expression of nitric oxide signaling enzymes in the injured developing lung and in vascular smooth muscle cells.Am J Physiol Lung Cell Mol Physiol. 2010 Mar;298(3):L324-34. doi: 10.1152/ajplung.00181.2009. Epub 2009 Dec 18. Am J Physiol Lung Cell Mol Physiol. 2010. PMID: 20023176 Free PMC article.
-
Nitric oxide regulation of fetal and newborn lung development and function.Nitric Oxide. 2024 Jun 1;147:13-25. doi: 10.1016/j.niox.2024.04.005. Epub 2024 Apr 7. Nitric Oxide. 2024. PMID: 38588917 Free PMC article. Review.
-
Soluble guanylate cyclase modulates alveolarization in the newborn lung.Am J Physiol Lung Cell Mol Physiol. 2013 Oct 15;305(8):L569-81. doi: 10.1152/ajplung.00401.2012. Epub 2013 Aug 9. Am J Physiol Lung Cell Mol Physiol. 2013. PMID: 23934926 Free PMC article.
References
-
- Lincoln TM, Dey N, Sellak H. Invited review: cGMP-dependent protein kinase signaling mechanisms in smooth muscle: from the regulation of tone to gene expression. J Appl Physiol. 2001;91:1421–30. - PubMed
-
- Pilz RB, Broderick KE. Role of cyclic GMP in gene regulation. Front Biosci. 2005;10:1239–68. - PubMed
-
- Gudi T, Casteel DE, Vinson C, Boss GR, Pilz RB. NO activation of fos promoter elements requires nuclear translocation of G-kinase I and CREB phosphorylation but is independent of MAP kinase activation. Oncogene. 2000;19:6324–33. - PubMed
-
- Gudi T, Huvar I, Meinecke M, Lohmann SM, Boss GR, Pilz RB. Regulation of gene expression by cGMP-dependent protein kinase. Transactivation of the c-fos promoter. J Biol Chem. 1996;271:4597–600. - PubMed
-
- Sauzeau V, Rolli-Derkinderen M, Marionneau C, Loirand G, Pacaud P. RhoA expression is controlled by nitric oxide through cGMP-dependent protein kinase activation. J Biol Chem. 2003;278:9472–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

