Genetic identification of a network of factors that functionally interact with the nucleosome remodeling ATPase ISWI
- PMID: 18535655
- PMCID: PMC2390755
- DOI: 10.1371/journal.pgen.1000089
Genetic identification of a network of factors that functionally interact with the nucleosome remodeling ATPase ISWI
Abstract
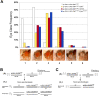
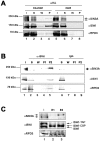
Nucleosome remodeling and covalent modifications of histones play fundamental roles in chromatin structure and function. However, much remains to be learned about how the action of ATP-dependent chromatin remodeling factors and histone-modifying enzymes is coordinated to modulate chromatin organization and transcription. The evolutionarily conserved ATP-dependent chromatin-remodeling factor ISWI plays essential roles in chromosome organization, DNA replication, and transcription regulation. To gain insight into regulation and mechanism of action of ISWI, we conducted an unbiased genetic screen to identify factors with which it interacts in vivo. We found that ISWI interacts with a network of factors that escaped detection in previous biochemical analyses, including the Sin3A gene. The Sin3A protein and the histone deacetylase Rpd3 are part of a conserved histone deacetylase complex involved in transcriptional repression. ISWI and the Sin3A/Rpd3 complex co-localize at specific chromosome domains. Loss of ISWI activity causes a reduction in the binding of the Sin3A/Rpd3 complex to chromatin. Biochemical analysis showed that the ISWI physically interacts with the histone deacetylase activity of the Sin3A/Rpd3 complex. Consistent with these findings, the acetylation of histone H4 is altered when ISWI activity is perturbed in vivo. These findings suggest that ISWI associates with the Sin3A/Rpd3 complex to support its function in vivo.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev. 2003;13:136–142. - PubMed
-
- Iizuka M, Smith MM. Functional consequences of histone modifications. Curr Opin Genet Dev. 2003;13:154–160. - PubMed
-
- Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. - PubMed
-
- van Vugt JJ, Ranes M, Campsteijn C, Logie C. The ins and outs of ATP-dependent chromatin remodeling in budding yeast: biophysical and proteomic perspectives. Biochim Biophys Acta. 2007;1769:153–171. - PubMed
-
- Dirscherl SS, Krebs JE. Functional diversity of ISWI complexes. Biochem Cell Biol. 2004;82:482–489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

