Evolutionary expansion and anatomical specialization of synapse proteome complexity
- PMID: 18536710
- PMCID: PMC3624047
- DOI: 10.1038/nn.2135
Evolutionary expansion and anatomical specialization of synapse proteome complexity
Abstract
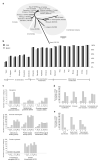
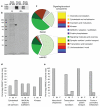
Understanding the origins and evolution of synapses may provide insight into species diversity and the organization of the brain. Using comparative proteomics and genomics, we examined the evolution of the postsynaptic density (PSD) and membrane-associated guanylate kinase (MAGUK)-associated signaling complexes (MASCs) that underlie learning and memory. PSD and MASC orthologs found in yeast carry out basic cellular functions to regulate protein synthesis and structural plasticity. We observed marked changes in signaling complexity at the yeast-metazoan and invertebrate-vertebrate boundaries, with an expansion of key synaptic components, notably receptors, adhesion/cytoskeletal proteins and scaffold proteins. A proteomic comparison of Drosophila and mouse MASCs revealed species-specific adaptation with greater signaling complexity in mouse. Although synaptic components were conserved amongst diverse vertebrate species, mapping mRNA and protein expression in the mouse brain showed that vertebrate-specific components preferentially contributed to differences between brain regions. We propose that the evolution of synapse complexity around a core proto-synapse has contributed to invertebrate-vertebrate differences and to brain specialization.
Figures


References
-
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–8. - PubMed
-
- Moore BR. The evolution of learning. Biol Rev Camb Philos Soc. 2004;79:301–35. - PubMed
-
- Roth G, Dicke U. Evolution of the brain and intelligence. Trends Cogn Sci. 2005;9:250–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

