The gastric HK-ATPase: structure, function, and inhibition
- PMID: 18536934
- PMCID: PMC3079481
- DOI: 10.1007/s00424-008-0495-4
The gastric HK-ATPase: structure, function, and inhibition
Erratum in
- Pflugers Arch. 2011 Mar;461(3):399
Abstract
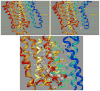
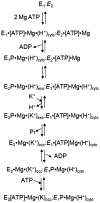
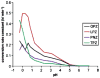
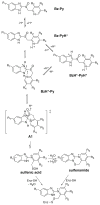
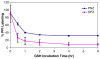
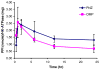
The gastric H,K-ATPase, a member of the P(2)-type ATPase family, is the integral membrane protein responsible for gastric acid secretion. It is an alpha,beta-heterodimeric enzyme that exchanges cytoplasmic hydronium with extracellular potassium. The catalytic alpha subunit has ten transmembrane segments with a cluster of intramembranal carboxylic amino acids located in the middle of the transmembrane segments TM4, TM5,TM6, and TM8. Comparison to the known structure of the SERCA pump, mutagenesis, and molecular modeling has identified these as constituents of the ion binding domain. The beta subunit has one transmembrane segment with N terminus in cytoplasmic region. The extracellular domain of the beta subunit contains six or seven N-linked glycosylation sites. N-glycosylation is important for the enzyme assembly, maturation, and sorting. The enzyme pumps acid by a series of conformational changes from an E(1) (ion site in) to an E(2) (ion site out) configuration following binding of MgATP and phosphorylation. Several experimental observations support the hypothesis that expulsion of the proton at 160 mM (pH 0.8) results from movement of lysine 791 into the ion binding site in the E(2)P configuration. Potassium access from the lumen depends on activation of a K and Cl conductance via a KCNQ1/KCNE2 complex and Clic6. K movement through the luminal channel in E(2)P is proposed to displace the lysine along with dephosphorylation to return the enzyme to the E(1) configuration. This enzyme is inhibited by the unique proton pump inhibitor class of drug, allowing therapy of acid-related diseases.
Figures



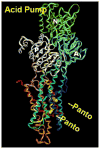
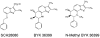
References
-
- Abe K, Kaya S, Taniguchi K, Hayashi Y, Imagawa T, Kikumoto M, Oiwa K, Sakaguchi K. Evidence for a relationship between activity and the tetraprotomeric assembly of solubilized pig gastric H/K-ATPase. J Biochem (Tokyo) 2005;138:293–301. - PubMed
-
- Andersson K, Carlsson E. Potassium-competitive acid blockade: a new therapeutic strategy in acid-related diseases. Pharmacol Ther. 2005;108:294–307. - PubMed
-
- Asano S, Yoshida A, Yashiro H, Kobayashi Y, Morisato A, Ogawa H, Takeguchi N, Morii M. The cavity structure for docking the K(+)-competitive inhibitors in the gastric proton pump. J Biol Chem. 2004;279:13968–13975. - PubMed
-
- Bamberg K, Mercier F, Reuben MA, Kobayashi Y, Munson KB, Sachs G. cDNA cloning and membrane topology of the rabbit gastric H+/K(+)-ATPase alpha-subunit. Biochim Biophys Acta. 1992;1131:69–77. - PubMed
-
- Besancon M, Shin JM, Mercier F, Munson K, Miller M, Hersey S, Sachs G. Membrane topology and omeprazole labeling of the gastric H+,K(+)-adenosinetriphosphatase. Biochemistry. 1993;32:2345–2355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

