Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia
- PMID: 18538732
- PMCID: PMC2486365
- DOI: 10.1016/j.ccr.2008.04.020
Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia
Abstract
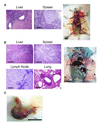
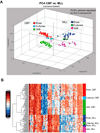
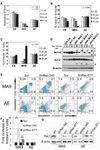
Faithful modeling of mixed-lineage leukemia in murine cells has been difficult to achieve. We show that expression of MLL-AF9 in human CD34+ cells induces acute myeloid, lymphoid, or mixed-lineage leukemia in immunodeficient mice. Some leukemia stem cells (LSC) were multipotent and could be lineage directed by altering either the growth factors or the recipient strain of mouse, highlighting the importance of microenvironmental cues. Other LSC were strictly lineage committed, demonstrating the heterogeneity of the stem cell compartment in MLL disease. Targeting the Rac signaling pathway by pharmacologic or genetic means resulted in rapid and specific apoptosis of MLL-AF9 cells, suggesting that the Rac signaling pathway may be a valid therapeutic target in MLL-rearranged AML.
Figures




Comment in
-
MLL-AF9 leukemia stem cells: hardwired or taking cues from the microenvironment?Cancer Cell. 2008 Jun;13(6):465-7. doi: 10.1016/j.ccr.2008.05.012. Cancer Cell. 2008. PMID: 18538728
References
-
- Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, Pieters R, Kersey JH, Sallan SE, Fletcher JA, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. - PubMed
-
- Armstrong SA, Mabon ME, Silverman LB, Li A, Gribben JG, Fox EA, Sallan SE, Korsmeyer SJ. FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103:3544–3546. - PubMed
-
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. - PubMed
-
- Baer MR, Stewart CC, Lawrence D, Arthur DC, Mrozek K, Strout MP, Davey FR, Schiffer CA, Bloomfield CD. Acute myeloid leukemia with 11q23 translocations: myelomonocytic immunophenotype by multiparameter flow cytometry. Leukemia. 1998;12:317–325. - PubMed
-
- Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

