Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization
- PMID: 18539308
- PMCID: PMC2516379
- DOI: 10.1016/j.virol.2008.04.045
Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization
Abstract
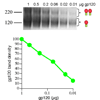
Insights into the process of HIV-1 neutralization may assist rational vaccine design. Here, we compared antibody neutralization against the JR-FL primary isolate and trimer binding affinities judged by native PAGE. Monovalent Fab-trimer binding and neutralization showed a direct quantitative relationship, implying that neutralization begins as each trimer is occupied by one antibody. At saturation, three Fab or soluble CD4 molecules engaged each trimer. In contrast, a maximum of one soluble CD4 molecule bound to functional SIV trimers with a truncated a gp41 tail. Remarkably, soluble CD4 was found to trigger dramatic enhancement of this virus. Unlike Fabs, a quantitative correlation between JR-FL trimer binding and neutralization was unclear for some, but not all IgGs, as neutralization was markedly increased, but trimer affinity was largely unchanged. In addition, only one molecule of certain gp41-specific IgGs appeared to be able to bind each trimer. We discuss the implications of these findings in weighing the relative contributions of size, multivalent binding and other possible effects of IgGs to explain their increased potency.
Figures




References
-
- Arthos J, Cicala C, Steenbeke TD, Chun TW, Dela Cruz C, Hanback DB, Khazanie P, Nam D, Schuck P, Selig SM, Van Ryk D, Chaikin MA, Fauci AS. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: implications for therapeutic and vaccine strategies. J Biol Chem. 2002;277(13):11456–11464. - PubMed
-
- Barbas CF, 3rd, Collet TA, Amberg W, Roben P, Binley JM, Hoekstra D, Cababa D, Jones TM, Williamson RA, Pilkington GR, et al. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J Mol Biol. 1993;230(3):812–823. - PubMed
-
- Bennett A, Liu J, Van Ryk D, Bliss D, Arthos J, Henderson RM, Subramaniam S. Cryoelectron tomographic analysis of an HIV-neutralizing protein and its complex with native viral gp120. J Biol Chem. 2007;282(38):27754–27759. - PubMed
-
- Binley JM, Ditzel HJ, Barbas CF, 3rd, Sullivan N, Sodroski J, Parren PW, Burton DR. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retroviruses. 1996;12(10):911–924. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

