ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system
- PMID: 18539740
- PMCID: PMC2493248
- DOI: 10.1128/JB.00321-08
ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system
Abstract
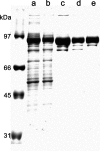
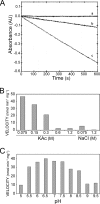
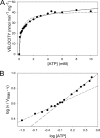
Type IV secretion systems (T4SS) mediate the transfer of DNA and protein substrates to target cells. TrwK, encoded by the conjugative plasmid R388, is a member of the VirB4 family, comprising the largest and most conserved proteins of T4SS. VirB4 was suggested to be an ATPase involved in energizing pilus assembly and substrate transport. However, conflicting experimental evidence concerning VirB4 ATP hydrolase activity was reported. Here, we demonstrate that TrwK is able to hydrolyze ATP in vitro in the absence of its potential macromolecular substrates and other T4SS components. The kinetic parameters of its ATPase activity have been characterized. The TrwK oligomerization state was investigated by analytical ultracentrifugation and electron microscopy, and its effects on ATPase activity were analyzed. The results suggest that the hexameric form of TrwK is the catalytically active state, much like the structurally related protein TrwB, the conjugative coupling protein.
Figures



References
-
- Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 431359-1365. - PubMed
-
- Batchelor, R. A., B. M. Pearson, L. M. Friis, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 1503507-3517. - PubMed
-
- Beijersbergen, A., S. J. Smith, and P. J. Hooykaas. 1994. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid 32212-218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

