SEL, a selective enrichment broth for simultaneous growth of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes
- PMID: 18539786
- PMCID: PMC2519329
- DOI: 10.1128/AEM.02756-07
SEL, a selective enrichment broth for simultaneous growth of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes
Abstract
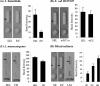
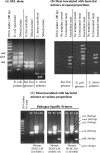
Multipathogen detection on a single-assay platform not only reduces the cost for testing but also provides data on the presence of pathogens in a single experiment. To achieve this detection, a multipathogen selective enrichment medium is essential to allow the concurrent growth of pathogens. SEL broth was formulated to allow the simultaneous growth of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes. The results were compared to those obtained with the respective individual selective enrichment broths, Rappaport-Vassiliadis (RV) for S. enterica, modified E. coli broth with 20 mg of novobiocin/liter for E. coli O157:H7, and Fraser broth for L. monocytogenes, and a currently used universal preenrichment broth (UPB). The growth of each pathogen in SEL inoculated at 10(1) or 10(3) CFU/ml was superior to that in the respective individual enrichment broth, except in the case of RV, in which Salmonella cells inoculated at both concentrations grew equally well. In mixed-culture experiments with cells of the three species present in equal concentrations or at a 1:10:1,000 ratio, the overall growth was proportional to the initial inoculation levels; however, the growth of L. monocytogenes was markedly suppressed when cells of this species were present at lower concentrations than those of the other two species. Further, SEL was able to resuscitate acid- and cold-stressed cells, and recovery was comparable to that in nonselective tryptic soy broth containing 6% yeast extract but superior to that in the respective individual selective broths. SEL promoted the growth of all three pathogens in a mixture in ready-to-eat salami and in turkey meat samples. Moreover, each pathogen was readily detected by a pathogen-specific immunochromatographic lateral-flow or multiplex PCR assay. Even though the growth of each pathogen in SEL was comparable to that in UPB, SEL inhibited greater numbers of nontarget organisms than did UPB. In summary, SEL was demonstrated to be a promising new multiplex selective enrichment broth for the detection of the three most prominent food-borne pathogens by antibody- or nucleic acid-based methods.
Figures

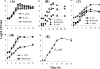

References
-
- Bailey, J. S., and N. A. Cox. 1992. Universal preenrichment broth for the simultaneous detection of Salmonella and Listeria in foods. J. Food Prot. 55:256-259. - PubMed
-
- Bhagwat, A. A. 2006. Microbiological safety of fresh-cut produce: where are we now?, p. 121-165. In K. R. Mathews (ed.), Microbiology of fresh produce. ASM Press, Washington, DC.
-
- Bhagwat, A. A. 2003. Simultaneous detection of Escherichia coli O157:H7, Listeria monocytogenes and Salmonella strains by real-time PCR. Int. J. Food Microbiol. 84:217-224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

