Comparative genomics of Lbx loci reveals conservation of identical Lbx ohnologs in bony vertebrates
- PMID: 18541024
- PMCID: PMC2446394
- DOI: 10.1186/1471-2148-8-171
Comparative genomics of Lbx loci reveals conservation of identical Lbx ohnologs in bony vertebrates
Abstract
Background: Lbx/ladybird genes originated as part of the metazoan cluster of Nk homeobox genes. In all animals investigated so far, both the protostome genes and the vertebrate Lbx1 genes were found to play crucial roles in neural and muscle development. Recently however, additional Lbx genes with divergent expression patterns were discovered in amniotes. Early in the evolution of vertebrates, two rounds of whole genome duplication are thought to have occurred, during which 4 Lbx genes were generated. Which of these genes were maintained in extant vertebrates, and how these genes and their functions evolved, is not known.
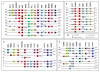
Results: Here we searched vertebrate genomes for Lbx genes and discovered novel members of this gene family. We also identified signature genes linked to particular Lbx loci and traced the remnants of 4 Lbx paralogons (two of which retain Lbx genes) in amniotes. In teleosts, that have undergone an additional genome duplication, 8 Lbx paralogons (three of which retain Lbx genes) were found. Phylogenetic analyses of Lbx and Lbx-associated genes show that in extant, bony vertebrates only Lbx1- and Lbx2-type genes are maintained. Of these, some Lbx2 sequences evolved faster and were probably subject to neofunctionalisation, while Lbx1 genes may have retained more features of the ancestral Lbx gene. Genes at Lbx1 and former Lbx4 loci are more closely related, as are genes at Lbx2 and former Lbx3 loci. This suggests that during the second vertebrate genome duplication, Lbx1/4 and Lbx2/3 paralogons were generated from the duplicated Lbx loci created during the first duplication event.
Conclusion: Our study establishes for the first time the evolutionary history of Lbx genes in bony vertebrates, including the order of gene duplication events, gene loss and phylogenetic relationships. Moreover, we identified genetic hallmarks for each of the Lbx paralogons that can be used to trace Lbx genes as other vertebrate genomes become available. Significantly, we show that bony vertebrates only retained copies of Lbx1 and Lbx2 genes, with some Lbx2 genes being highly divergent. Thus, we have established a base on which the evolution of Lbx gene function in vertebrate development can be evaluated.
Figures


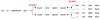
Similar articles
-
Conservation of gene linkage in dispersed vertebrate NK homeobox clusters.Dev Genes Evol. 2009 Oct;219(9-10):481-96. doi: 10.1007/s00427-009-0311-y. Dev Genes Evol. 2009. PMID: 20112453
-
Novel developmental bases for the evolution of hypobranchial muscles in vertebrates.BMC Biol. 2020 Sep 9;18(1):120. doi: 10.1186/s12915-020-00851-y. BMC Biol. 2020. PMID: 32907560 Free PMC article.
-
Comparative genomics of ParaHox clusters of teleost fishes: gene cluster breakup and the retention of gene sets following whole genome duplications.BMC Genomics. 2007 Sep 6;8:312. doi: 10.1186/1471-2164-8-312. BMC Genomics. 2007. PMID: 17822543 Free PMC article.
-
Evolution of endothelin receptors in vertebrates.Gen Comp Endocrinol. 2014 Dec 1;209:21-34. doi: 10.1016/j.ygcen.2014.06.028. Epub 2014 Jul 8. Gen Comp Endocrinol. 2014. PMID: 25010382 Review.
-
Diversification of four human HOX gene clusters by step-wise evolution rather than ancient whole-genome duplications.Dev Genes Evol. 2015 Nov;225(6):353-7. doi: 10.1007/s00427-015-0518-z. Epub 2015 Oct 19. Dev Genes Evol. 2015. PMID: 26481129 Review.
Cited by
-
O-GlcNAc cycling: emerging roles in development and epigenetics.Semin Cell Dev Biol. 2010 Aug;21(6):646-54. doi: 10.1016/j.semcdb.2010.05.001. Epub 2010 May 19. Semin Cell Dev Biol. 2010. PMID: 20488252 Free PMC article. Review.
-
Unravelling paralogous gene expression dynamics during three-spined stickleback embryogenesis.Sci Rep. 2019 Mar 6;9(1):3752. doi: 10.1038/s41598-019-40127-2. Sci Rep. 2019. PMID: 30842559 Free PMC article.
-
Role of Primary Cilia in Skeletal Disorders.Stem Cells Int. 2022 Jun 18;2022:6063423. doi: 10.1155/2022/6063423. eCollection 2022. Stem Cells Int. 2022. PMID: 35761830 Free PMC article. Review.
-
Lbx2 regulates formation of myofibrils.BMC Dev Biol. 2009 Feb 12;9:13. doi: 10.1186/1471-213X-9-13. BMC Dev Biol. 2009. PMID: 19216761 Free PMC article.
-
Role of zebrafish lbx2 in embryonic lateral line development.PLoS One. 2011;6(12):e29515. doi: 10.1371/journal.pone.0029515. Epub 2011 Dec 22. PLoS One. 2011. PMID: 22216300 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

