Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae
- PMID: 18541657
- PMCID: PMC2493207
- DOI: 10.1128/IAI.01613-07
Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae
Abstract
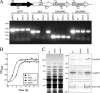
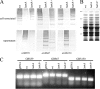
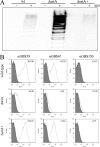
Pili are putative virulence factors and promising vaccine candidates in Streptococcus agalactiae (group B Streptococcus [GBS]) infection, a leading cause of neonatal sepsis and meningitis. The genes necessary for pilus synthesis and assembly are clustered in pilus islands (PI). Each gene encodes three structural subunits (a backbone and two ancillary proteins) bearing a C-terminal LPXTG motif and two subfamily C sortases (SrtC) involved in covalent polymerization of the subunits. GBS strains also possess the conserved "housekeeping" sortase A (SrtA), but its role in pilus assembly is unclear. To address this issue, pilus expression and cell wall anchoring were analyzed in srtA deletion mutants. Loss of SrtA did not affect pilus polymerization. However, pilus expression on the cell surface was reduced, and pili accumulated in the culture supernatant. Furthermore, cell-associated pili could be readily released by detergent treatment, indicating that SrtA is involved in covalent anchoring of pili to the cell wall. When each of the genes comprising PI-2a was systematically deleted, only the absence of ancillary subunit GBS150 or the SrtC required for incorporation of GBS150 into pili mimicked the srtA mutant phenotype. Thus, from these data a model for GBS pilus assembly can be proposed in which PI sortases are responsible for polymerization of the pilus structure, while SrtA is required to covalently attach it to the cell wall, utilizing ancillary pilus subunit GBS150 as the anchor protein.
Figures
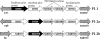


Similar articles
-
Sortase A substrate specificity in GBS pilus 2a cell wall anchoring.PLoS One. 2011;6(10):e25300. doi: 10.1371/journal.pone.0025300. Epub 2011 Oct 4. PLoS One. 2011. PMID: 21991306 Free PMC article.
-
Structural differences between the Streptococcus agalactiae housekeeping and pilus-specific sortases: SrtA and SrtC1.PLoS One. 2011;6(8):e22995. doi: 10.1371/journal.pone.0022995. Epub 2011 Aug 30. PLoS One. 2011. PMID: 21912586 Free PMC article.
-
Roles of the sortases of Streptococcus pneumoniae in assembly of the RlrA pilus.J Bacteriol. 2008 Sep;190(17):6002-13. doi: 10.1128/JB.00379-08. Epub 2008 Jul 7. J Bacteriol. 2008. PMID: 18606733 Free PMC article.
-
Pilus biogenesis of Gram-positive bacteria: Roles of sortases and implications for assembly.Protein Sci. 2017 Aug;26(8):1458-1473. doi: 10.1002/pro.3191. Epub 2017 May 15. Protein Sci. 2017. PMID: 28493331 Free PMC article. Review.
-
Pili with strong attachments: Gram-positive bacteria do it differently.Mol Microbiol. 2006 Oct;62(2):320-30. doi: 10.1111/j.1365-2958.2006.05279.x. Epub 2006 Sep 15. Mol Microbiol. 2006. PMID: 16978260 Review.
Cited by
-
Dynamics of fecal microbial communities in children with diarrhea of unknown etiology and genomic analysis of associated Streptococcus lutetiensis.BMC Microbiol. 2013 Jun 19;13:141. doi: 10.1186/1471-2180-13-141. BMC Microbiol. 2013. PMID: 23782707 Free PMC article.
-
Preliminary crystallographic study of the Streptococcus agalactiae sortases, sortase A and sortase C1.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Sep 1;66(Pt 9):1096-100. doi: 10.1107/S1744309110031106. Epub 2010 Aug 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20823536 Free PMC article.
-
Sortase enzymes in Gram-positive bacteria.Mol Microbiol. 2011 Dec;82(5):1044-59. doi: 10.1111/j.1365-2958.2011.07887.x. Epub 2011 Nov 7. Mol Microbiol. 2011. PMID: 22026821 Free PMC article. Review.
-
Streptococcus agalactiae Non-Pilus, Cell Wall-Anchored Proteins: Involvement in Colonization and Pathogenesis and Potential as Vaccine Candidates.Front Immunol. 2018 Apr 5;9:602. doi: 10.3389/fimmu.2018.00602. eCollection 2018. Front Immunol. 2018. PMID: 29686667 Free PMC article. Review.
-
Increase in pilus islet 2-encoded pili among Streptococcus pneumoniae isolates, Atlanta, Georgia, USA.Emerg Infect Dis. 2010 Jun;16(6):955-62. doi: 10.3201/eid1606.091820. Emerg Infect Dis. 2010. PMID: 20507746 Free PMC article.
References
-
- Abbot, E. L., W. D. Smith, G. P. S. Siou, C. Chiriboga, R. J. Smith, J. A. Wilson, B. H. Hirst, and M. A. Kehoe. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 91822-1833. - PubMed
-
- Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 1032857-2862. - PMC - PubMed
-
- Budzik, J. M., L. A. Marraffini, and O. Schneewind. 2007. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 66495-510. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

