Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36
- PMID: 18541663
- PMCID: PMC2519698
- DOI: 10.1128/MCB.00001-08
Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36
Abstract
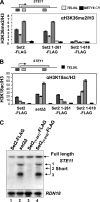
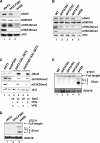
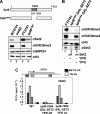
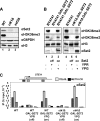
Set2 (KMT3)-dependent methylation (me) of histone H3 at lysine 36 (H3K36) promotes deacetylation of transcribed chromatin and represses cryptic promoters within genes. Although Set2 is the only methyltransferase (KMTase) for H3K36 in yeast, it is not known if Set2 is regulated or whether the different methylation states at H3K36 are functionally distinct. Here we show that the N-terminal 261 residues of Set2 (Set2(1-261)), containing the SET KMTase domain, are sufficient for H3K36me2, histone deacetylation, and repression of cryptic promoters at STE11. Set2-catalyzed H3K36me2 does not require either Ctk1-dependent phosphorylation of RNA polymerase II (RNAPII) or the presence of the phospho-C-terminal domain (CTD) interaction (SRI) domain of Set2. This finding is consistent with a known correlation between H3K36me2 and whether a gene is on or off, but not the level of activity of a gene. By contrast, H3K36me3 requires Spt6, proline 38 on histone H3 (H3P38), the CTD of RNAPII, Ctk1, and the C-terminal SRI domain of Set2. We suggest that the C-terminal region of Set2, in conjunction with the phosphorylated CTD of RNAPII, influences the KMTase activity to promote H3K36me3 during transcription elongation.
Figures



References
-
- Adkins, M. W., and J. K. Tyler. 2006. Transcriptional activators are dispensable for transcription in the absence of spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21405-416. - PubMed
-
- Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 2721473-1476. - PubMed
-
- Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123581-592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
