The cis-acting replication elements define human enterovirus and rhinovirus species
- PMID: 18541697
- PMCID: PMC2491478
- DOI: 10.1261/rna.1031408
The cis-acting replication elements define human enterovirus and rhinovirus species
Abstract
Replication of picornaviruses is dependent on VPg uridylylation, which is linked to the presence of the internal cis-acting replication element (cre). Cre are located within the sequence encoding polyprotein, yet at distinct positions as demonstrated for poliovirus and coxsackievirus-B3, cardiovirus, and human rhinovirus (HRV-A and HRV-B), overlapping proteins 2C, VP2, 2A, and VP1, respectively. Here we report a novel distinct cre element located in the VP2 region of the recently reported HRV-A2 species and provide evolutionary evidence of its functionality. We also experimentally interrogated functionality of recently identified HRV-B cre in the 2C region that is orthologous to the human enterovirus (HEV) cre and show that it is dispensable for replication and appears to be a nonfunctional evolutionary relic. In addition, our mutational analysis highlights two amino acids in the 2C protein that are crucial for replication. Remarkably, we conclude that each genetic clade of HRV and HEV is characterized by a unique functional cre element, where evolutionary success of a new genetic lineage seems to be associated with an invention of a novel cre motif and decay of the ancestral one. Therefore, we propose that cre element could be considered as an additional criterion for human rhinovirus and enterovirus classification.
Figures


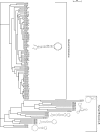

References
-
- Andino R., Rieckhof G.E., Baltimore D. A functional ribonucleoprotein complex forms around the 5′-end of poliovirus RNA. Cell. 1990;63:369–380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
