Centromere mitotic recombination in mammalian cells
- PMID: 18541703
- PMCID: PMC2426939
- DOI: 10.1083/jcb.200803042
Centromere mitotic recombination in mammalian cells
Abstract
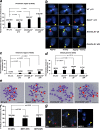
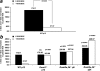
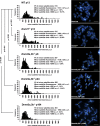
Centromeres are special structures of eukaryotic chromosomes that hold sister chromatid together and ensure proper chromosome segregation during cell division. Centromeres consist of repeated sequences, which have hindered the study of centromere mitotic recombination and its consequences for centromeric function. We use a chromosome orientation fluorescence in situ hybridization technique to visualize and quantify recombination events at mouse centromeres. We show that centromere mitotic recombination occurs in normal cells to a higher frequency than telomere recombination and to a much higher frequency than chromosome-arm recombination. Furthermore, we show that centromere mitotic recombination is increased in cells lacking the Dnmt3a and Dnmt3b DNA methyltransferases, suggesting that the epigenetic state of centromeric heterochromatin controls recombination events at these regions. Increased centromere recombination in Dnmt3a,3b-deficient cells is accompanied by changes in the length of centromere repeats, suggesting that prevention of illicit centromere recombination is important to maintain centromere integrity in the mouse.
Figures

References
-
- Bailey, S.M., E.H. Goodwin, J. Meyne, and M.N. Cornforth. 1996. CO-FISH reveals inversions associated with isochromosome formation. Mutagenesis. 11:139–144. - PubMed
-
- Bechter, O.E., J.W. Shay, and W.E. Wright. 2004. The frequency of homologous recombination in human ALT cells. Cell Cycle. 3:547–549. - PubMed
-
- Blackburn, E.H. 2001. Switching and signalling at the telomere. Cell. 106:661–673. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

