Specific DNA-binding by apicomplexan AP2 transcription factors
- PMID: 18541913
- PMCID: PMC2423414
- DOI: 10.1073/pnas.0801993105
Specific DNA-binding by apicomplexan AP2 transcription factors
Abstract
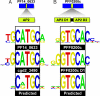
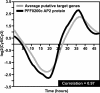
Malaria remains one of the most prevalent infectious diseases worldwide, affecting more than half a billion people annually. Despite many years of research, the mechanisms underlying transcriptional regulation in the malaria-causing Plasmodium spp., and in Apicomplexan parasites generally, remain poorly understood. In Plasmodium, few regulatory elements sufficient to drive gene expression have been characterized, and their cognate DNA-binding proteins remain unknown. This study characterizes the DNA-binding specificities of two members of the recently identified Apicomplexan AP2 (ApiAP2) family of putative transcriptional regulators from Plasmodium falciparum. The ApiAP2 proteins contain AP2 domains homologous to the well characterized plant AP2 family of transcriptional regulators, which play key roles in development and environmental stress response pathways. We assayed ApiAP2 protein-DNA interactions using protein-binding microarrays and combined these results with computational predictions of coexpressed target genes to couple these putative trans factors to corresponding cis-regulatory motifs in Plasmodium. Furthermore, we show that protein-DNA sequence specificity is conserved in orthologous proteins between phylogenetically distant Apicomplexan species. The identification of the DNA-binding specificities for ApiAP2 proteins lays the foundation for the exploration of their role as transcriptional regulators during all stages of parasite development. Because of their origin in the plant lineage, ApiAP2 proteins have no homologues in the human host and may prove to be ideal antimalarial targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Sachs J, Malaney P. The economic and social burden of malaria. Nature. 2002;415:680–685. - PubMed
-
- Hall N, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. - PubMed
-
- Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

