Mutations of the GABA-A receptor alpha1 subunit M1 domain reveal unexpected complexity for modulation by neuroactive steroids
- PMID: 18544665
- PMCID: PMC2701692
- DOI: 10.1124/mol.108.048520
Mutations of the GABA-A receptor alpha1 subunit M1 domain reveal unexpected complexity for modulation by neuroactive steroids
Abstract
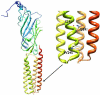
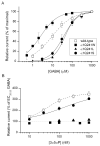
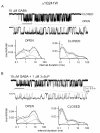
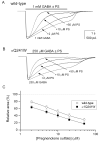
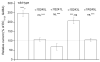
Neuroactive steroids are among the most efficacious modulators of the mammalian GABA-A receptor. Previous work has proposed that receptor potentiation is mediated by steroid interactions with a site defined by the residues alpha1Asn407/Tyr410 in the M4 transmembrane domain and residue alpha1Gln241 in the M1 domain. We examined the role of residues in the alpha1 subunit M1 domain in the modulation of the rat alpha1beta2gamma2L GABA-A receptor by neuroactive steroids. The data demonstrate that the region is critical to the actions of potentiating neuroactive steroids. Receptors containing the alpha1Q241W or alpha1Q241L mutations were insensitive to (3alpha,5alpha)-3-hydroxypregnan-20-one (3alpha5alphaP), albeit with different underlying mechanisms. The alpha1Q241S mutant was potentiated by 3alpha5alphaP, but the kinetic mode of potentiation was altered by the mutation. It is noteworthy that the alpha1Q241L mutation had no effect on channel potentiation by (3alpha,5alpha)-3-hydroxymethyl-pregnan-20-one, but mutation of the neighboring residue, alpha1Ser240, prevented channel modulation. A steroid lacking an H-bonding group on C3 (5alpha-pregnan-20-one) potentiated the wild-type receptor but not the alpha1Q241L mutant. The findings are consistent with a model in which the alpha1Ser240 and alpha1Gln241 residues shape the surface to which steroid molecules bind.
Figures

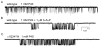





References
-
- Boileau AJ, Li T, Benkwitz C, Czajkowski C, Pearce RA. Effects of the γ2S subunit incorporation on GABAA receptor macroscopic kinetics. Neuropharmacol. 2003;44:1003–1012. - PubMed
-
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

