Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny
- PMID: 18545663
- PMCID: PMC2398773
- DOI: 10.1371/journal.pone.0002388
Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny
Abstract
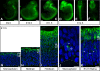
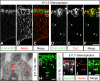
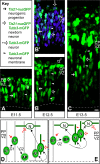
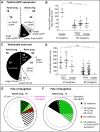
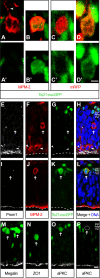
The neurons of the mammalian brain are generated by progenitors dividing either at the apical surface of the ventricular zone (neuroepithelial and radial glial cells, collectively referred to as apical progenitors) or at its basal side (basal progenitors, also called intermediate progenitors). For apical progenitors, the orientation of the cleavage plane relative to their apical-basal axis is thought to be of critical importance for the fate of the daughter cells. For basal progenitors, the relationship between cell polarity, cleavage plane orientation and the fate of daughter cells is unknown. Here, we have investigated these issues at the very onset of cortical neurogenesis. To directly observe the generation of neurons from apical and basal progenitors, we established a novel transgenic mouse line in which membrane GFP is expressed from the beta-III-tubulin promoter, an early pan-neuronal marker, and crossed this line with a previously described knock-in line in which nuclear GFP is expressed from the Tis21 promoter, a pan-neurogenic progenitor marker. Mitotic Tis21-positive basal progenitors nearly always divided symmetrically, generating two neurons, but, in contrast to symmetrically dividing apical progenitors, lacked apical-basal polarity and showed a nearly randomized cleavage plane orientation. Moreover, the appearance of beta-III-tubulin-driven GFP fluorescence in basal progenitor-derived neurons, in contrast to that in apical progenitor-derived neurons, was so rapid that it suggested the initiation of the neuronal phenotype already in the progenitor. Our observations imply that (i) the loss of apical-basal polarity restricts neuronal progenitors to the symmetric mode of cell division, and that (ii) basal progenitors initiate the expression of neuronal phenotype already before mitosis, in contrast to apical progenitors.
Conflict of interest statement
Figures


References
-
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. - PubMed
-
- McConnell SK. Constructing the cerebral cortex: neurogenesis and fate determination. Neuron. 1995;15:761–768. - PubMed
-
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. - PubMed
-
- Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

