Gremlin enhances the determined path to cardiomyogenesis
- PMID: 18545679
- PMCID: PMC2398777
- DOI: 10.1371/journal.pone.0002407
Gremlin enhances the determined path to cardiomyogenesis
Abstract
Background: The critical event in heart formation is commitment of mesodermal cells to a cardiomyogenic fate, and cardiac fate determination is regulated by a series of cytokines. Bone morphogenetic proteins (BMPs) and fibroblast growth factors have been shown to be involved in this process, however additional factors needs to be identified for the fate determination, especially at the early stage of cardiomyogenic development.
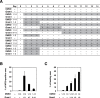
Methodology/principal findings: Global gene expression analysis using a series of human cells with a cardiomyogenic potential suggested Gremlin (Grem1) is a candidate gene responsible for in vitro cardiomyogenic differentiation. Grem1, a known BMP antagonist, enhanced DMSO-induced cardiomyogenesis of P19CL6 embryonal carcinoma cells (CL6 cells) 10-35 fold in an area of beating differentiated cardiomyocytes. The Grem1 action was most effective at the early differentiation stage when CL6 cells were destined to cardiomyogenesis, and was mediated through inhibition of BMP2. Furthermore, BMP2 inhibited Wnt/beta-catenin signaling that promoted CL6 cardiomyogenesis.
Conclusions/significance: Grem1 enhances the determined path to cardiomyogenesis in a stage-specific manner, and inhibition of the BMP signaling pathway is involved in initial determination of Grem1-promoted cardiomyogenesis. Our results shed new light on renewal of the cardiovascular system using Grem1 in human.
Conflict of interest statement
Figures




References
-
- Andree B, Duprez D, Vorbusch B, Arnold HH, Brand T. BMP-2 induces ectopic expression of cardiac lineage markers and interferes with somite formation in chicken embryos. Mech Dev. 1998;70:119–131. - PubMed
-
- Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11:451–462. - PubMed
-
- Alsan BH, Schultheiss TM. Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002;129:1935–1943. - PubMed
-
- Crossley PH, Martin GR. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

