Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors
- PMID: 18547390
- PMCID: PMC2610386
- DOI: 10.1111/j.1365-2958.2008.06301.x
Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors
Abstract
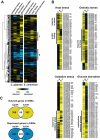
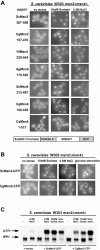
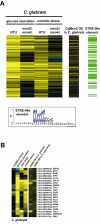
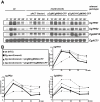
We determined the genome-wide environmental stress response (ESR) expression profile of Candida glabrata, a human pathogen related to Saccharomyces cerevisiae. Despite different habitats, C. glabrata, S. cerevisiae, Schizosaccharomyces pombe and Candida albicans have a qualitatively similar ESR. We investigate the function of the C. glabrata syntenic orthologues to the ESR transcription factor Msn2. The C. glabrata orthologues CgMsn2 and CgMsn4 contain a motif previously referred to as HD1 (homology domain 1) also present in Msn2 orthologues from fungi closely related to S. cerevisiae. We show that regions including this motif confer stress-regulated intracellular localization when expressed in S. cerevisiae. Site-directed mutagenesis confirms that nuclear export of CgMsn2 in C. glabrata requires an intact HD1. Transcript profiles of CgMsn2/4 mutants and CgMsn2 overexpression strains show that they regulate a part of the CgESR. CgMsn2 complements a S. cerevisiae msn2 null mutant and in stressed C. glabrata cells, rapidly translocates from the cytosol to the nucleus. CgMsn2 is required for full resistance against severe osmotic stress and rapid and full induction of trehalose synthesis genes (TPS1, TPS2). Constitutive activation of CgMsn2 is detrimental for C. glabrata. These results establish an Msn2-regulated general stress response in C. glabrata.
Figures




References
-
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. - PubMed
-
- Bilsland E, Molin C, Swaminathan S, Ramne A, Sunnerhagen P. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol Microbiol. 2004;53:1743–1756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

