Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins
- PMID: 18547521
- PMCID: PMC2519881
- DOI: 10.1016/j.str.2008.03.009
Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins
Abstract
Ras GTPases are conformational switches controlling cell proliferation, differentiation, and development. Despite their prominent role in many forms of cancer, the mechanism of conformational transition between inactive GDP-bound and active GTP-bound states remains unclear. Here we describe a detailed analysis of available experimental structures and molecular dynamics simulations to quantitatively assess the structural and dynamical features of active and inactive states and their interconversion. We demonstrate that GTP-bound and nucleotide-free G12V H-ras sample a wide region of conformational space, and show that the inactive-to-active transition is a multiphase process defined by the relative rearrangement of the two switches and the orientation of Tyr32. We also modeled and simulated N- and K-ras proteins and found that K-ras is more flexible than N- and H-ras. We identified a number of isoform-specific, long-range side chain interactions that define unique pathways of communication between the nucleotide binding site and the C terminus.
Figures
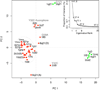


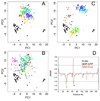


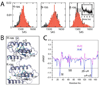


References
-
- Abseher R, Nilges M. Are there non-trivial dynamic cross-correlations in proteins? Journal of Molecular Biology. 1998;279:911–920. - PubMed
-
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

