RNA dynamics: it is about time
- PMID: 18547802
- PMCID: PMC2580758
- DOI: 10.1016/j.sbi.2008.04.004
RNA dynamics: it is about time
Abstract
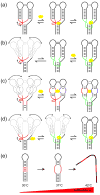
Many recently discovered RNA functions rely on highly complex multistep conformational transitions that occur in response to an array of cellular signals. These dynamics accompany and guide, for example, RNA cotranscriptional folding, ligand sensing and signaling, site-specific catalysis in ribozymes, and the hierarchically ordered assembly of ribonucleoproteins. RNA dynamics are encoded by both the inherent properties of RNA structure, spanning many motional modes with a large range of amplitudes and timescales, and external trigger factors, ranging from proteins, nucleic acids, metal ions, metabolites, and vitamins to temperature and even directional RNA biosynthesis itself. Here, we review recent advances in our understanding of RNA dynamics as highlighted by biophysical tools.
Figures
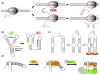

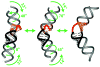
References
-
- Nagel JH, Pleij CW. Self-induced structural switches in RNA. Biochimie. 2002;84:913–923. - PubMed
-
- Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. - PubMed
-
- Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc Natl Acad Sci U S A. 2007;104:17995–18000. This paper shows for three different non-coding RNAs that their sequences code for transcription pause sites that facilitate folding of long helices during co-transcriptional folding. - PMC - PubMed
-
- Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

