Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA
- PMID: 18548002
- PMCID: PMC2856441
- DOI: 10.1038/nature07106
Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA
Abstract
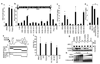
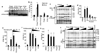
Innate immune defences are essential for the control of virus infection and are triggered through host recognition of viral macromolecular motifs known as pathogen-associated molecular patterns (PAMPs). Hepatitis C virus (HCV) is an RNA virus that replicates in the liver, and infects 200 million people worldwide. Infection is regulated by hepatic immune defences triggered by the cellular RIG-I helicase. RIG-I binds PAMP RNA and signals interferon regulatory factor 3 activation to induce the expression of interferon-alpha/beta and antiviral/interferon-stimulated genes (ISGs) that limit infection. Here we identify the polyuridine motif of the HCV genome 3' non-translated region and its replication intermediate as the PAMP substrate of RIG-I, and show that this and similar homopolyuridine or homopolyriboadenine motifs present in the genomes of RNA viruses are the chief feature of RIG-I recognition and immune triggering in human and murine cells. 5' terminal triphosphate on the PAMP RNA was necessary but not sufficient for RIG-I binding, which was primarily dependent on homopolymeric ribonucleotide composition, linear structure and length. The HCV PAMP RNA stimulated RIG-I-dependent signalling to induce a hepatic innate immune response in vivo, and triggered interferon and ISG expression to suppress HCV infection in vitro. These results provide a conceptual advance by defining specific homopolymeric RNA motifs within the genome of HCV and other RNA viruses as the PAMP substrate of RIG-I, and demonstrate immunogenic features of the PAMP-RIG-I interaction that could be used as an immune adjuvant for vaccine and immunotherapy approaches.
Figures


References
-
- Saito T, Gale M. Principles of intracellular viral recognition. Current Opinion in Immunology. 2007;19:17–23. - PubMed
-
- Lauer GM, Walker BD. Medical progress: Hepatitis C virus infection. New England Journal of Medicine. 2001;345:41–52. - PubMed
-
- Gale M, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. - PubMed
-
- Hornung V, et al. 5 '-triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

