Substrate-targeting gamma-secretase modulators
- PMID: 18548070
- PMCID: PMC2678541
- DOI: 10.1038/nature07055
Substrate-targeting gamma-secretase modulators
Abstract
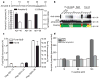
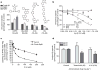
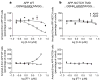
Selective lowering of Abeta42 levels (the 42-residue isoform of the amyloid-beta peptide) with small-molecule gamma-secretase modulators (GSMs), such as some non-steroidal anti-inflammatory drugs, is a promising therapeutic approach for Alzheimer's disease. To identify the target of these agents we developed biotinylated photoactivatable GSMs. GSM photoprobes did not label the core proteins of the gamma-secretase complex, but instead labelled the beta-amyloid precursor protein (APP), APP carboxy-terminal fragments and amyloid-beta peptide in human neuroglioma H4 cells. Substrate labelling was competed by other GSMs, and labelling of an APP gamma-secretase substrate was more efficient than a Notch substrate. GSM interaction was localized to residues 28-36 of amyloid-beta, a region critical for aggregation. We also demonstrate that compounds known to interact with this region of amyloid-beta act as GSMs, and some GSMs alter the production of cell-derived amyloid-beta oligomers. Furthermore, mutation of the GSM binding site in the APP alters the sensitivity of the substrate to GSMs. These findings indicate that substrate targeting by GSMs mechanistically links two therapeutic actions: alteration in Abeta42 production and inhibition of amyloid-beta aggregation, which may synergistically reduce amyloid-beta deposition in Alzheimer's disease. These data also demonstrate the existence and feasibility of 'substrate targeting' by small-molecule effectors of proteolytic enzymes, which if generally applicable may significantly broaden the current notion of 'druggable' targets.
Figures

Comment in
-
Biochemistry: Molecular cloaking devices.Nature. 2008 Jun 12;453(7197):861-2. doi: 10.1038/453861a. Nature. 2008. PMID: 18548057 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

