Genomic sequencing of single microbial cells from environmental samples
- PMID: 18550420
- PMCID: PMC3635501
- DOI: 10.1016/j.mib.2008.05.006
Genomic sequencing of single microbial cells from environmental samples
Abstract
Recently developed techniques allow genomic DNA sequencing from single microbial cells [Lasken RS: Single-cell genomic sequencing using multiple displacement amplification. Curr Opin Microbiol 2007, 10:510-516]. Here, we focus on research strategies for putting these methods into practice in the laboratory setting. An immediate consequence of single-cell sequencing is that it provides an alternative to culturing organisms as a prerequisite for genomic sequencing. The microgram amounts of DNA required as template are amplified from a single bacterium by a method called multiple displacement amplification (MDA) avoiding the need to grow cells. The ability to sequence DNA from individual cells will likely have an immense impact on microbiology considering the vast numbers of novel organisms, which have been inaccessible unless culture-independent methods could be used. However, special approaches have been necessary to work with amplified DNA. MDA may not recover the entire genome from the single copy present in most bacteria. Also, some sequence rearrangements can occur during the DNA amplification reaction. Over the past two years many research groups have begun to use MDA, and some practical approaches to single-cell sequencing have been developed. We review the consensus that is emerging on optimum methods, reliability of amplified template, and the proper interpretation of 'composite' genomes which result from the necessity of combining data from several single-cell MDA reactions in order to complete the assembly. Preferred laboratory methods are considered on the basis of experience at several large sequencing centers where >70% of genomes are now often recovered from single cells. Methods are reviewed for preparation of bacterial fractions from environmental samples, single-cell isolation, DNA amplification by MDA, and DNA sequencing.
Figures
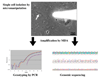
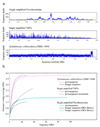
References
-
- Lasken RS. Single-cell genomic sequencing using Multiple Displacement Amplification. Curr Opin Microbiol. 2007;10:510–516. - PubMed
-
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. - PubMed
-
- Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial Ecology and Evolution: A Ribosomal RNA Approach. Annual Review of Microbiology. 1986;40:337–365. - PubMed
-
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

