Genome evolution of Wolbachia strain wPip from the Culex pipiens group
- PMID: 18550617
- PMCID: PMC2515876
- DOI: 10.1093/molbev/msn133
Genome evolution of Wolbachia strain wPip from the Culex pipiens group
Abstract
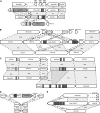
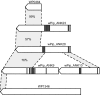
The obligate intracellular bacterium Wolbachia pipientis strain wPip induces cytoplasmic incompatibility (CI), patterns of crossing sterility, in the Culex pipiens group of mosquitoes. The complete sequence is presented of the 1.48-Mbp genome of wPip which encodes 1386 coding sequences (CDSs), representing the first genome sequence of a B-supergroup Wolbachia. Comparisons were made with the smaller genomes of Wolbachia strains wMel of Drosophila melanogaster, an A-supergroup Wolbachia that is also a CI inducer, and wBm, a mutualist of Brugia malayi nematodes that belongs to the D-supergroup of Wolbachia. Despite extensive gene order rearrangement, a core set of Wolbachia genes shared between the 3 genomes can be identified and contrasts with a flexible gene pool where rapid evolution has taken place. There are much more extensive prophage and ankyrin repeat encoding (ANK) gene components of the wPip genome compared with wMel and wBm, and both are likely to be of considerable importance in wPip biology. Five WO-B-like prophage regions are present and contain some genes that are identical or highly similar in multiple prophage copies, whereas other genes are unique, and it is likely that extensive recombination, duplication, and insertion have occurred between copies. A much larger number of genes encode ankyrin repeat (ANK) proteins in wPip, with 60 present compared with 23 in wMel, many of which are within or close to the prophage regions. It is likely that this pattern is partly a result of expansions in the wPip lineage, due for example to gene duplication, but their presence is in some cases more ancient. The wPip genome underlines the considerable evolutionary flexibility of Wolbachia, providing clear evidence for the rapid evolution of ANK-encoding genes and of prophage regions. This host-Wolbachia system, with its complex patterns of sterility induced between populations, now provides an excellent model for unraveling the molecular systems underlying host reproductive manipulation.
Figures

References
-
- Baldo L, Werren JH. Revisiting Wolbachia supergroup typing based on WSP: spurious lineages and discordance with MLST. Curr Microbiol. 2007;55:81–87. - PubMed
-
- Bandi C, McCall JW, Genchi C, Corona S, Venco L, Sacchi L. Effects of tetracycline on the filarial worms Brugia pahangi and Dirofilaria immitis and their bacterial endosymbionts Wolbachia. Int J Parasitol. 1999;29:357–364. - PubMed
-
- Barr AR. Cytoplasmic incompatibility in natural populations of a mosquito, Culex pipiens L. Nature. 1980;283:71–72. - PubMed
-
- Bordenstein SR, Wernegreen JJ. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 2004;21:1981–1991. - PubMed
-
- Brownlie JC, O'Neill SL. Wolbachia genomics: accelerating our understanding of a pervasive symbiosis. In: Bourtzis K, Miller TA, editors. Insect symbiosis. Boca Raton (FL): CRC Press; 2006. pp. 175–183.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

