Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya
- PMID: 18550667
- PMCID: PMC2519552
- DOI: 10.1128/JVI.01115-08
Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya
Abstract
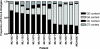
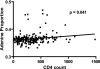
APOBEC3G is an important innate immune molecule that causes human immunodeficiency virus type 1 (HIV-1) hypermutation, which can result in detrimental viral genome mutations. The Vif protein of wild-type HIV-1 counteracts APOBEC3G activity by targeting it for degradation and inhibiting its incorporation into viral particles. Additional APOBEC cytidine deaminases have been identified, such as APOBEC3F, which has a similar mode of action but different sequence specificity. A relationship between APOBEC3F/G and HIV disease progression has been proposed. During HIV-1 sequence analysis of the vpu/env region of 240 HIV-infected subjects from Nairobi, Kenya, 13 drastically hypermutated proviral sequences were identified. Sequences derived from plasma virus, however, lacked hypermutation, as did proviral vif. When correlates of disease progression were examined, subjects with hypermutated provirus were found to have significantly higher CD4 counts than the other subjects. Furthermore, hypermutation as estimated by elevated adenine content positively correlated with CD4 count for all 240 study subjects. The sequence context of the observed hypermutation was statistically associated with APOBEC3F/G activity. In contrast to previous studies, this study demonstrates that higher CD4 counts correlate with increased hypermutation in the absence of obvious mutations in the APOBEC inhibiting Vif protein. This strongly suggests that host factors, such as APOBEC3F/G, are playing a protective role in these patients, modulating viral hypermutation and host disease progression. These findings support the potential of targeting APOBEC3F/G for therapeutic purposes.
Figures

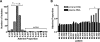


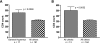
References
-
- Biasin, M., L. Piacentini, C. S. Lo, Y. Kanari, G. Magri, D. Trabattoni, V. Naddeo, L. Lopalco, A. Clivio, E. Cesana, F. Fasano, C. Bergamaschi, F. Mazzotta, M. Miyazawa, and M. Clerici. 2007. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G: a possible role in the resistance to HIV of HIV-exposed seronegative individuals. J. Infect. Dis. 195960-964. - PubMed
-
- Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 141392-1396. - PubMed
-
- Bonvin, M., F. Achermann, I. Greeve, D. Stroka, A. Keogh, D. Inderbitzin, D. Candinas, P. Sommer, S. Wain-Hobson, J. P. Vartanian, and J. Greeve. 2006. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 431364-1374. - PubMed
-
- Braddick, M. R., J. K. Kreiss, J. B. Embree, P. Datta, J. O. Ndinya-Achola, H. Pamba, G. Maitha, P. L. Roberts, T. C. Quinn, and K. K. Holmes. 1990. Impact of maternal HIV infection on obstetrical and early neonatal outcome. AIDS 41001-1005. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

