Quantitative 1H nuclear magnetic resonance metabolite profiling as a functional genomics platform to investigate alkaloid biosynthesis in opium poppy
- PMID: 18550684
- PMCID: PMC2492654
- DOI: 10.1104/pp.108.120493
Quantitative 1H nuclear magnetic resonance metabolite profiling as a functional genomics platform to investigate alkaloid biosynthesis in opium poppy
Abstract
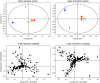
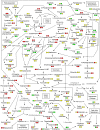
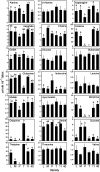
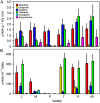
Opium poppy (Papaver somniferum) produces a diverse array of bioactive benzylisoquinoline alkaloids and has emerged as a versatile model system to study plant alkaloid metabolism. The plant is widely cultivated as the only commercial source of the narcotic analgesics morphine and codeine. Variations in plant secondary metabolism as a result of genetic diversity are often associated with perturbations in other metabolic pathways. As part of a functional genomics platform, we used (1)H nuclear magnetic resonance (NMR) metabolite profiling for the analysis of primary and secondary metabolism in opium poppy. Aqueous and chloroform extracts of six different opium poppy cultivars were subjected to chemometric analysis. Principle component analysis of the (1)H NMR spectra for latex extracts clearly distinguished two varieties, including a low-alkaloid variety and a high-thebaine, low-morphine cultivar. Distinction was also made between pharmaceutical-grade opium poppy cultivars and a condiment variety. Such phenotypic differences were not observed in root extracts. Loading plots confirmed that morphinan alkaloids contributed predominantly to the variance in latex extracts. Quantification of 34 root and 21 latex metabolites, performed using Chenomx NMR Suite version 4.6, showed major differences in the accumulation of specific alkaloids in the latex of the low-alkaloid and high-thebaine, low-morphine varieties. Relatively few differences were found in the levels of other metabolites, indicating that the variation was specific for alkaloid metabolism. Exceptions in the low-alkaloid cultivar included an increased accumulation of the alkaloid precursor tyramine and reduced levels of sucrose, some amino acids, and malate. Real-time polymerase chain reaction analysis of 42 genes involved in primary and secondary metabolism showed differential gene expression mainly associated with alkaloid biosynthesis. Reduced alkaloid levels in the condiment variety were associated with the reduced abundance of transcripts encoding several alkaloid biosynthetic enzymes.
Figures



References
-
- Allen RS, Miller JAC, Chitty JA, Fist AJ, Gerlach WL, Larkin PJ (2008) Metabolic engineering of morphinan alkaloids by over-expression and RNAi suppression of salutaridinol 7-O-acetyltransferase in opium poppy. Plant Biotechnol J 6 22–30 - PubMed
-
- Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JAC, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22 1559–1566 - PubMed
-
- Baker JM, Hawkins ND, Ward JL, Lovegrove A, Napier JA, Shewry PR, Beale MH (2006) A metabolomic study of substantial equivalence of field-grown genetically modified wheat. Plant Biotechnol J 4 381–392 - PubMed
-
- Bernáth J, Németh É, Petheõ F (2003) Alkaloid accumulation in capsules of the selfed and cross-pollinated poppy. Plant Breed 122 263–267
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

