Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant
- PMID: 18550696
- PMCID: PMC2658752
- DOI: 10.1136/jmg.2007.055202
Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant
Abstract
Background: Genomic disorders are often caused by non-allelic homologous recombination between segmental duplications. Chromosome 16 is especially rich in a chromosome-specific low copy repeat, termed LCR16.
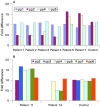
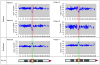
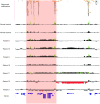
Methods and results: A bacterial artificial chromosome (BAC) array comparative genome hybridisation (CGH) screen of 1027 patients with mental retardation and/or multiple congenital anomalies (MR/MCA) was performed. The BAC array CGH screen identified five patients with deletions and five with apparently reciprocal duplications of 16p13 covering 1.65 Mb, including 15 RefSeq genes. In addition, three atypical rearrangements overlapping or flanking this region were found. Fine mapping by high-resolution oligonucleotide arrays suggests that these deletions and duplications result from non-allelic homologous recombination (NAHR) between distinct LCR16 subunits with >99% sequence identity. Deletions and duplications were either de novo or inherited from unaffected parents. To determine whether these imbalances are associated with the MR/MCA phenotype or whether they might be benign variants, a population of 2014 normal controls was screened. The absence of deletions in the control population showed that 16p13.11 deletions are significantly associated with MR/MCA (p = 0.0048). Despite phenotypic variability, common features were identified: three patients with deletions presented with MR, microcephaly and epilepsy (two of these had also short stature), and two other deletion carriers ascertained prenatally presented with cleft lip and midline defects. In contrast to its previous association with autism, the duplication seems to be a common variant in the population (5/1682, 0.29%).
Conclusion: These findings indicate that deletions inherited from clinically normal parents are likely to be causal for the patients' phenotype whereas the role of duplications (de novo or inherited) in the phenotype remains uncertain. This difference in knowledge regarding the clinical relevance of the deletion and the duplication causes a paradigm shift in (cyto)genetic counselling.
Conflict of interest statement
Figures

References
-
- Inoue K, Lupski JR. Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 2002;3:199–242 - PubMed
-
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science 2002;297:1003–7 - PubMed
-
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, Fitzpatrick CA, Segraves R, Richmond TA, Guiver C, Albertson DG, Pinkel D, Eis PS, Schwartz S, Knight SJ, Eichler EE. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet 2006;38:1038–42 - PubMed
-
- Balciuniene J, Feng N, Iyadurai K, Hirsch B, Charnas L, Bill BR, Easterday MC, Staaf J, Oseth L, Czapansky-Beilman D, Avramopoulos D, Thomas GH, Borg A, Valle D, Schimmenti LA, Selleck SB. Recurrent 10q22–q23 deletions: a genomic disorder on 10q associated with cognitive and behavioral abnormalities. Am J Hum Genet 2007;80:938–47 - PMC - PubMed
-
- Loftus BJ, Kim UJ, Sneddon VP, Kalush F, Brandon R, Fuhrmann J, Mason T, Crosby ML, Barnstead M, Cronin L, Deslattes Mays A, Cao Y, Xu RX, Kang HL, Mitchell S, Eichler EE, Harris PC, Venter JC, Adams MD. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics 1999;60:295–308 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
