A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome
- PMID: 18550805
- PMCID: PMC2493396
- DOI: 10.1101/gr.078261.108
A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome
Abstract
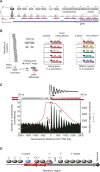
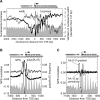
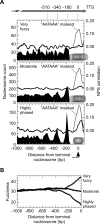
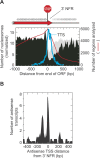
Most nucleosomes are well-organized at the 5' ends of S. cerevisiae genes where "-1" and "+1" nucleosomes bracket a nucleosome-free promoter region (NFR). How nucleosomal organization is specified by the genome is less clear. Here we establish and inter-relate rules governing genomic nucleosome organization by sequencing DNA from more than one million immunopurified S. cerevisiae nucleosomes (displayed at http://atlas.bx.psu.edu/). Evidence is presented that the organization of nucleosomes throughout genes is largely a consequence of statistical packing principles. The genomic sequence specifies the location of the -1 and +1 nucleosomes. The +1 nucleosome forms a barrier against which nucleosomes are packed, resulting in uniform positioning, which decays at farther distances from the barrier. We present evidence for a novel 3' NFR that is present at >95% of all genes. 3' NFRs may be important for transcription termination and anti-sense initiation. We present a high-resolution genome-wide map of TFIIB locations that implicates 3' NFRs in gene looping.
Figures


References
-
- Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. - PubMed
-
- Alen C., Kent N.A., Jones H.S., O'Sullivan J., Aranda A., Proudfoot N.J. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell. 2002;10:1441–1452. - PubMed
-
- Bao Y., White C.L., Luger K. Nucleosome core particles containing a poly(dA⋅dT) sequence element exhibit a locally distorted DNA structure. J. Mol. Biol. 2006;361:617–624. - PubMed
-
- Basehoar A.D., Zanton S.J., Pugh B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
