Comparing proteins by their unfolding pattern
- PMID: 18550806
- PMCID: PMC2426622
- DOI: 10.1529/biophysj.108.129999
Comparing proteins by their unfolding pattern
Abstract
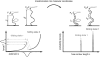
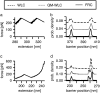
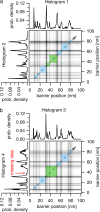
Single molecule force spectroscopy has evolved into an important and extremely powerful technique for investigating the folding potentials of biomolecules. Mechanical tension is applied to individual molecules, and the subsequent, often stepwise unfolding is recorded in force extension traces. However, because the energy barriers of the folding potentials are often close to the thermal energy, both the extensions and the forces at which these barriers are overcome are subject to marked fluctuations. Therefore, force extension traces are an inadequate representation despite widespread use particularly when large populations of proteins need to be compared and analyzed. We show in this article that contour length, which is independent of fluctuations and alterable experimental parameters, is a more appropriate variable than extension. By transforming force extension traces into contour length space, histograms are obtained that directly represent the energy barriers. In contrast to force extension traces, such barrier position histograms can be averaged to investigate details of the unfolding potential. The cross-superposition of barrier position histograms allows us to detect and visualize the order of unfolding events. We show with this approach that in contrast to the sequential unfolding of bacteriorhodopsin, two main steps in the unfolding of the enzyme titin kinase are independent of each other. The potential of this new method for accurate and automated analysis of force spectroscopy data and for novel automated screening techniques is shown with bacteriorhodopsin and with protein constructs containing GFP and titin kinase.
Figures
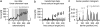



References
-
- Binnig, G., C. F. Quate, and C. Gerber. 1986. Atomic force microscope. Phys. Rev. Lett. 56:930–933. - PubMed
-
- Block, S. M., L. S. Goldstein, and B. J. Schnapp. 1990. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 348:348–352. - PubMed
-
- Smith, S. B., Y. J. Cui, and C. Bustamante. 1996. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 271:795–799. - PubMed
-
- Smith, S. B., L. Finzi, and C. Bustamante. 1992. Direct mechanical measurements of the elasticity of single DNA-molecules by using magnetic beads. Science. 258:1122–1126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

