Effect of glycosylation on protein folding: a close look at thermodynamic stabilization
- PMID: 18550810
- PMCID: PMC2448824
- DOI: 10.1073/pnas.0801340105
Effect of glycosylation on protein folding: a close look at thermodynamic stabilization
Abstract
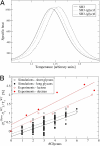
Glycosylation is one of the most common posttranslational modifications to occur in protein biosynthesis, yet its effect on the thermodynamics and kinetics of proteins is poorly understood. A minimalist model based on the native protein topology, in which each amino acid and sugar ring was represented by a single bead, was used to study the effect of glycosylation on protein folding. We studied in silico the folding of 63 engineered SH3 domain variants that had been glycosylated with different numbers of conjugated polysaccharide chains at different sites on the protein's surface. Thermal stabilization of the protein by the polysaccharide chains was observed in proportion to the number of attached chains. Consistent with recent experimental data, the degree of thermal stabilization depended on the position of the glycosylation sites, but only very weakly on the size of the glycans. A thermodynamic analysis showed that the origin of the enhanced protein stabilization by glycosylation is destabilization of the unfolded state rather than stabilization of the folded state. The higher free energy of the unfolded state is enthalpic in origin because the bulky polysaccharide chains force the unfolded ensemble to adopt more extended conformations by prohibiting formation of a residual structure. The thermodynamic stabilization induced by glycosylation is coupled with kinetic stabilization. The effects introduced by the glycans on the biophysical properties of proteins are likely to be relevant to other protein polymeric conjugate systems that regularly occur in the cell as posttranslational modifications or for biotechnological purposes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

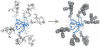


References
-
- Fersht AR, Daggett V. Protein folding and unfolding at atomic resolution. Cell. 2002;108:573–582. - PubMed
-
- Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38:245–288. - PubMed
-
- Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. - PubMed
-
- Jin W, Kambara O, Sasakawa H, Tamura A, Takada S. De novo design of foldable proteins with smooth folding funnel: Automated negative design and experimental verification. Structure. 2003;11:581–590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

