Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly
- PMID: 18550837
- PMCID: PMC2426963
- DOI: 10.1073/pnas.0803330105
Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly
Abstract
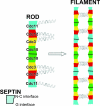
Mitotic yeast cells express five septins (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1/Sep7). Only Shs1 is nonessential. The four essential septins form a complex containing two copies of each, but their arrangement was not known. Single-particle analysis by EM confirmed that the heterooligomer is octameric and revealed that the subunits are arrayed in a linear rod. Identity of each subunit was determined by examining complexes lacking a given septin, by antibody decoration, and by fusion to marker proteins (GFP or maltose binding protein). The rod has the order Cdc11-Cdc12-Cdc3-Cdc10-Cdc10-Cdc3-Cdc12-Cdc11 and, hence, lacks polarity. At low ionic strength, rods assemble end-to-end to form filaments but not when Cdc11 is absent or its N terminus is altered. Filaments invariably pair into long parallel "railroad tracks." Lateral association seems to be mediated by heterotetrameric coiled coils between the paired C-terminal extensions of Cdc3 and Cdc12 projecting orthogonally from each filament. Shs1 may be able to replace Cdc11 at the end of the rod. Our findings provide insights into the molecular mechanisms underlying the function and regulation of cellular septin structures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. - PubMed
-
- Harbury PB, et al. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

