Overlapping protein-encoding genes in Pseudomonas fluorescens Pf0-1
- PMID: 18551168
- PMCID: PMC2396522
- DOI: 10.1371/journal.pgen.1000094
Overlapping protein-encoding genes in Pseudomonas fluorescens Pf0-1
Abstract
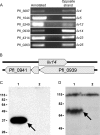
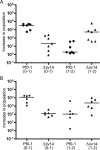
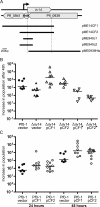
The annotated genome sequences of prokaryotes seldom include overlapping genes encoded opposite each other by the same stretch of DNA. However, antisense transcription is becoming recognized as a widespread phenomenon in eukaryotes, and examples have been linked to important biological processes. Pseudomonas fluorescens inhabits aquatic and terrestrial environments, and can be regarded as an environmental generalist. The genetic basis for this ecological success is not well understood. In a previous search for soil-induced genes in P. fluorescens Pf0-1, ten antisense genes were discovered. These were termed 'cryptic' genes, as they had escaped detection by gene-hunting algorithms, and lacked easily recognizable promoters. In this communication, we designate such genes as 'non-predicted' or 'hidden'. Using reverse transcription PCR, we show that at each of six non-predicted gene loci chosen for study, transcription occurs from both 'sense' and 'antisense' DNA strands. Further, at least one of these hidden antisense genes, iiv14, encodes a protein, as does the sense transcript, both identified by poly-histidine tags on the C-terminus of the proteins. Mutational and complementation studies showed that this novel antisense gene was important for efficient colonization of soil, and multiple copies in the wildtype host improved the speed of soil colonization. Introduction of a stop codon early in the gene eliminated complementation, further implicating the protein in colonization of soil. We therefore designate iiv14 "cosA". These data suggest that, as is the case with eukaryotes, some bacterial genomes are more densely coded than currently recognized.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Colonization strategies of Pseudomonas fluorescens Pf0-1: activation of soil-specific genes important for diverse and specific environments.BMC Microbiol. 2013 Apr 27;13:92. doi: 10.1186/1471-2180-13-92. BMC Microbiol. 2013. PMID: 23622502 Free PMC article.
-
Increased fitness of Pseudomonas fluorescens Pf0-1 leucine auxotrophs in soil.Appl Environ Microbiol. 2008 Jun;74(12):3644-51. doi: 10.1128/AEM.00429-08. Epub 2008 Apr 25. Appl Environ Microbiol. 2008. PMID: 18441116 Free PMC article.
-
Requirement of polyphosphate by Pseudomonas fluorescens Pf0-1 for competitive fitness and heat tolerance in laboratory media and sterile soil.Appl Environ Microbiol. 2009 Jun;75(12):3872-81. doi: 10.1128/AEM.00017-09. Epub 2009 Apr 24. Appl Environ Microbiol. 2009. PMID: 19395572 Free PMC article.
-
Use of in vivo expression technology to identify genes important in growth and survival of Pseudomonas fluorescens Pf0-1 in soil: discovery of expressed sequences with novel genetic organization.J Bacteriol. 2004 Nov;186(21):7411-9. doi: 10.1128/JB.186.21.7411-7419.2004. J Bacteriol. 2004. PMID: 15489453 Free PMC article.
-
[Overlapping genes and antisense transcription in eukaryotes].Genetika. 2014 Jul;50(7):749-64. Genetika. 2014. PMID: 25720133 Review. Russian.
Cited by
-
Colonization strategies of Pseudomonas fluorescens Pf0-1: activation of soil-specific genes important for diverse and specific environments.BMC Microbiol. 2013 Apr 27;13:92. doi: 10.1186/1471-2180-13-92. BMC Microbiol. 2013. PMID: 23622502 Free PMC article.
-
Regulation of polyphosphate kinase production by antisense RNA in Pseudomonas fluorescens Pf0-1.Appl Environ Microbiol. 2012 Jun;78(12):4533-7. doi: 10.1128/AEM.07836-11. Epub 2012 Apr 6. Appl Environ Microbiol. 2012. PMID: 22492458 Free PMC article.
-
Spotlight on alternative frame coding: Two long overlapping genes in Pseudomonas aeruginosa are translated and under purifying selection.iScience. 2022 Feb 1;25(2):103844. doi: 10.1016/j.isci.2022.103844. eCollection 2022 Feb 18. iScience. 2022. PMID: 35198897 Free PMC article.
-
Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens.Genome Biol. 2009;10(5):R51. doi: 10.1186/gb-2009-10-5-r51. Epub 2009 May 11. Genome Biol. 2009. PMID: 19432983 Free PMC article.
-
Bacterial antisense RNAs: how many are there, and what are they doing?Annu Rev Genet. 2010;44:167-88. doi: 10.1146/annurev-genet-102209-163523. Annu Rev Genet. 2010. PMID: 20707673 Free PMC article. Review.
References
-
- Rainey PB. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ Microbiol. 1999;1:243–257. - PubMed
-
- Brown DG, Allen C. Ralstonia solanacearum genes induced during growth in tomato: an inside view of bacterial wilt. Mol Microbiol. 2004;53:1641–1660. - PubMed
-
- Gal M, Preston GM, Massey RC, Spiers AJ, Rainey PB. Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol Ecol. 2003;12:3109–3121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

