Application of association mapping to understanding the genetic diversity of plant germplasm resources
- PMID: 18551188
- PMCID: PMC2423417
- DOI: 10.1155/2008/574927
Application of association mapping to understanding the genetic diversity of plant germplasm resources
Abstract
Compared to the conventional linkage mapping, linkage disequilibrium (LD)-mapping, using the nonrandom associations of loci in haplotypes, is a powerful high-resolution mapping tool for complex quantitative traits. The recent advances in the development of unbiased association mapping approaches for plant population with their successful applications in dissecting a number of simple to complex traits in many crop species demonstrate a flourish of the approach as a "powerful gene tagging" tool for crops in the plant genomics era of 21st century. The goal of this review is to provide nonexpert readers of crop breeding community with (1) the basic concept, merits, and simple description of existing methodologies for an association mapping with the recent improvements for plant populations, and (2) the details of some of pioneer and recent studies on association mapping in various crop species to demonstrate the feasibility, success, problems, and future perspectives of the efforts in plants. This should be helpful for interested readers of international plant research community as a guideline for the basic understanding, choosing the appropriate methods, and its application.
Figures
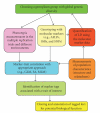
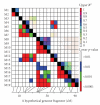

References
-
- Bermejo JEH, Leon J, editors. Neglected Crops 1492 from a Different Perspective. Botanical Garden of Córdoba, Andalusia, Spain: Food and Agriculture Organization (FAO) Corporate Document Repository; 1992. http://www.fao.org/docrep/T0646E/T0646E01.htm.
-
- Food and Health Organization's 1999 Report on the State of Food Insecurity in the World, http://www.fao.org/News/1999/img/SOFI99-E.PDF.
-
- Van Esbroeck GA, Bowman DT, May OL, Calhoun DS. Genetic similarity indices for ancestral cotton cultivars and their impact on genetic diversity estimates of modern cultivars. Crop Science. 1999;39(2):323–328.
-
- Meilleur BA, Hodgkin T. In situ conservation of crop wild relatives: status and trends. Biodiversity and Conservation. 2004;13(4):663–684.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

