Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model
- PMID: 18551192
- PMCID: PMC2423864
- DOI: 10.1172/JCI34753
Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model
Abstract
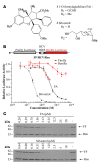
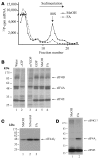
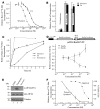
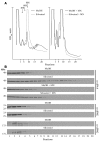
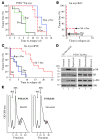
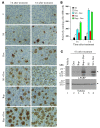
Disablement of cell death programs in cancer cells contributes to drug resistance and in some cases has been associated with altered translational control. As eukaryotic translation initiation factor 4E (eIF4E) cooperates with c-Myc during lymphomagenesis, induces drug resistance, and is a genetic modifier of the rapamycin response, we have investigated the effect of dysregulation of the ribosome recruitment phase of translation initiation on tumor progression and chemosensitivity. eIF4E is a subunit of eIF4F, a complex that stimulates ribosome recruitment during translation initiation by delivering the DEAD-box RNA helicase eIF4A to the 5' end of mRNAs. eIF4A is thought to prepare a ribosome landing pad on mRNA templates for incoming 40S ribosomes (and associated factors). Using small molecule screening, we found that cyclopenta[b]benzofuran flavaglines, a class of natural products, modulate eIF4A activity and inhibit translation initiation. One member of this class of compounds, silvestrol, was able to enhance chemosensitivity in a mouse lymphoma model in which carcinogenesis is driven by phosphatase and tensin homolog (PTEN) inactivation or elevated eIF4E levels. These results establish that targeting translation initiation can restore drug sensitivity in vivo and provide an approach to modulating chemosensitivity.
Figures
References
-
- Raught, B., Gingras, A.-C., and Sonenberg, N. 2000.Regulation of ribosome recruitment in eukaryotes. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 245–293.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

