Using GenePattern for gene expression analysis
- PMID: 18551415
- PMCID: PMC3893799
- DOI: 10.1002/0471250953.bi0712s22
Using GenePattern for gene expression analysis
Abstract
The abundance of genomic data now available in biomedical research has stimulated the development of sophisticated statistical methods for interpreting the data, and of special visualization tools for displaying the results in a concise and meaningful manner. However, biologists often find these methods and tools difficult to understand and use correctly. GenePattern is a freely available software package that addresses this issue by providing more than 100 analysis and visualization tools for genomic research in a comprehensive user-friendly environment for users at all levels of computational experience and sophistication. This unit demonstrates how to prepare and analyze microarray data in GenePattern.
Copyright 2008 by John Wiley & Sons, Inc.
Figures
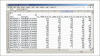

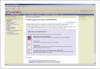
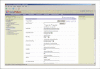
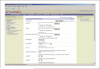
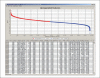

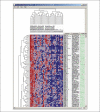


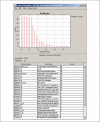
References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300.
-
- Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. Wadsworth & Brooks/Cole Advanced Books & Software; Monterey, Calif: 1984.
-
- Cover TM, Hart PE. Nearest neighbor pattern classification. IEEE Trans. Info. Theory. 1967;13:21–27.
-
- D'haeseleer P. How does gene expression clustering work? Nat. Biotechnol. 2005;23:1499–1501. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

